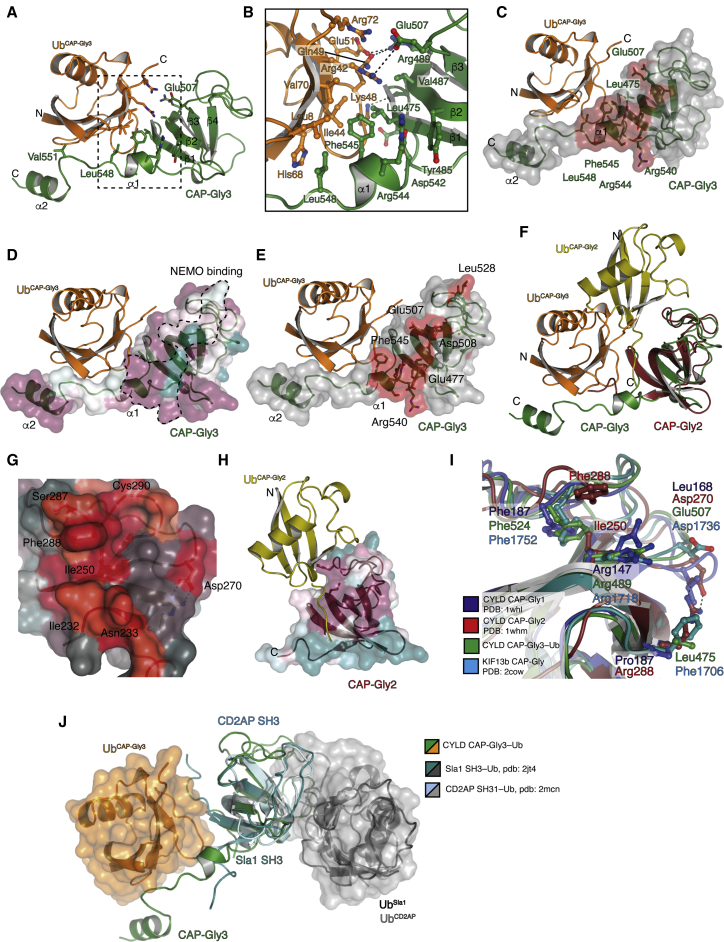
Figure 2.
CAP-Gly domains 2 and 3 of CYLD bind Ub
(A) Crystal structure of CAP-Gly3 (aa 464–565, green) to Ub (orange).
(B) Close-up view of the CAP-Gly3-Ub interface from (A). Interacting residues are shown as sticks.
(C) Surface of CAP-Gly3 (gray) with chemical shift perturbations (red) confirming the interaction interface is the same in solution.
(D) Surface view of CAP-Gly3 bound to Ub with residues colored onto the CAP-Gly3 surface based on conservation among different CYLD orthologs. Conservation scores were calculated with the Consurf server (Landau et al., 2005); purple, most conserved; blue, least conserved. Region interacting with NEMO, mapped by NMR in (E), is shown as a dotted line.
(E) Surface of CAP-Gly3 (gray) with chemical shift perturbations (red) of the interaction of CAP-Gly3 with NEMO ZnF.
(F) Superimposition of the CAP-Gly3-Ub structure onto an NMR-derived HADDOCK model of CAP-Gly2 binding to Ub (red and yellow, cartoon respectively), revealing an offset Ub-binding site for the two CAP-Gly domains.
(G) Surface of CAP-Gly2 (gray) with chemical shift perturbations (red) revealing the offset Ub-binding interface.
(H) Surface view of CAP-Gly2, colored by conservation as in (D), revealing that the Ub-interaction surface is conserved and offset to CAP-Gly3.
(I) Superimposition of the three CAP-Gly domains of CYLD: CAP-Gly1 (aa 120–213, PDB: 1whl, blue), CAP-Gly2-Ub (aa 226–313, PDB: 1whm, red), CAP-Gly3-Ub (aa 467–565, green), and KIF13b (aa 1685–1771, PDB: 2cow, cyan). Residues that coordinate Ub from the two different CAP-Gly2 and CAP-Gly3 interfaces are shown.
(J) Superimposition of SH3-Ub structures from Sla1 (PDB: 2jt4) and Cd2ap (PDB: 2mcn) onto CAP-Gly3, revealing the different modes of Ub binding by SH3 domains and CYLD CAP-Gly3.