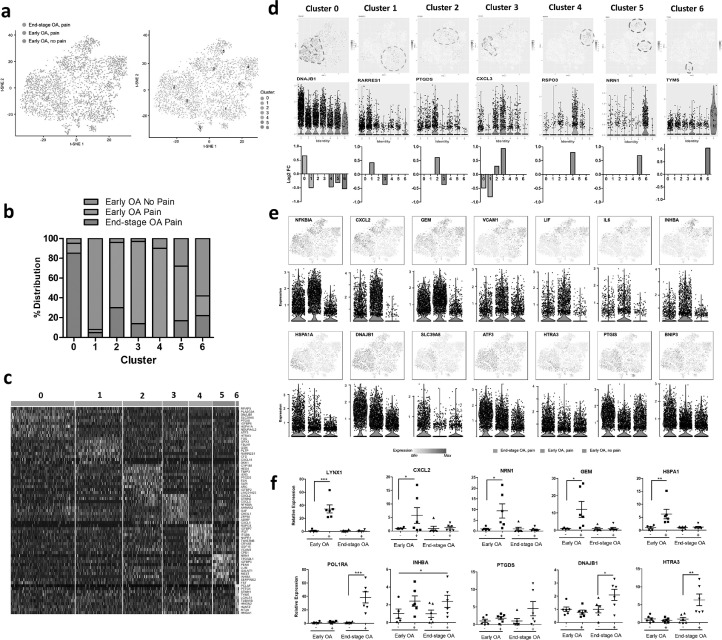
Fig. 3.
Single cell RNA-sequencing identifies distinct synovial fibroblasts subsets in the synovium from painful and non-painful sites in early OA and in end stage OA patients. (a) T-distributed stochastic neighbour embedding (t-SNE) plots of scRNAseq of synovial fibroblasts showing the separation between fibroblasts from painful and non-painful synovial sites in early OA patients, and between early and end-stage OA painful sites, with identification of 7 fibroblast subsets based on transcript profile. In total, scRNAseq was performed on synovial fibroblasts from painful and non-painful synovial patient-matched sites of n=4 early OA patients, and on synovial fibroblasts from painful sites of n = 4 end-stage OA patients. (b) Percentage distribution of the 7 different subsets according to either early OA pain, early OA no pain or end-stage OA pain. (c) Heatmap showing the z-score average gene signature expression of the top 10 most differentially expressed genes within each of the 7 synovial fibroblast clusters. (d) FeaturePlots displaying expression of subset specific markers for each of the 7 subsets (clusters 0-6) on the t-SNE map with violin plots below showing the expression levels of these markers (y-axis) across the different subsets (x-axis). (e) FeaturePlots displaying expression of sample specific markers (identified by performing differential expression analysis on the non-parametric Wilcoxon rank sum test) on the t-SNE map with violin plots below showing the expression levels of these markers (y-axis) across the different sample cohorts, early OA pain, early OA no-pain or end-stage OA pain (x-axis). (f) Bulk expression of specific synovial fibroblast genes in OA synovial tissue from either early OA patient matched painful (+) and non-painful (-) sites (n = 6) or end-stage OA patient-matched painful (+) and non-painful (-) sites (n = 6). *= p < 0.05, **= p < 0.01, ***= p < 0.001 significantly different between painful and non-painful synovial sites as determined by ANOVA with Tukey post-hoc test.