Table 2.
Analysis results of previously identified susceptibility loci. For the listed eight loci, MTOP global association test P-value was more than 10-fold lower compared to the standard logistic regression P-value. All of the loci are significant in global heterogeneity test (P 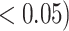 .
.
| SNP |
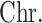
|
Position |
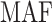
|
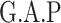
|
Standard analysis P |
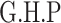
|
|---|---|---|---|---|---|---|
| rs4973768 | 3 | 27 416 013 | 0.47 | 3.1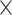 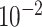
|
9.5 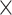 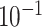
|
9.5 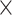 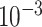
|
| rs10816625 | 9 | 110 837 073 | 0.06 | 5.0 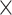 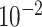
|
9.8 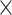 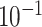
|
2.2 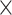 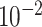
|
| rs7904519 | 10 | 114 773 927 | 0.46 | 6.5 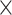 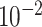
|
8.5 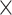 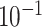
|
3.1 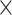 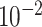
|
| rs554219 | 11 | 69 331 642 | 0.13 | 7.3 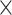 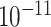
|
1.4 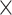 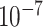
|
5.1 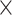 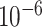
|
| rs11820646 | 11 | 129 461 171 | 0.40 | 1.5 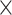 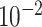
|
8.6 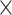 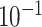
|
4.5 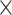 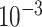
|
| rs2236007 | 14 | 37 132 769 | 0.21 | 2.1 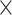 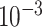
|
1.9 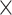 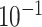
|
3.5 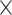 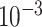
|
| rs1436904 | 18 | 24 570 667 | 0.40 | 7.2 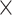 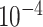
|
6.6 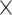 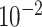
|
9.7 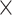 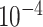
|
| rs1436904 | 22 | 29 121 087 | 0.01 | 9.8 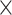 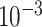
|
1.6 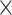 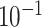
|
2.3 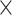 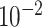
|
Chr. = chromosome; MAF = minor allele frequency; G.A.P = global association test P-value from MTOP; G.H.P = global heterogeneity test P-value from MTOP.
