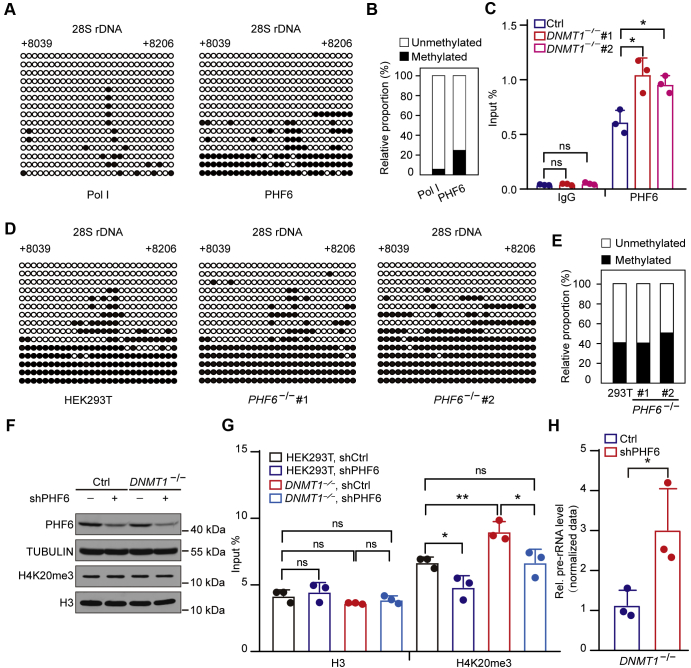
Figure 5.
PHF6 mediates the DNMT1-dependent inhibition of H4K20me3.A, PHF6 associates with both methylated and unmethylated DNA, and the latter account for more proportion. Pol I was used as a control, which binds exclusively to unmethylated CpG sites. ChIP assays, followed by bisulfite sequencing, showing the methylation status of DNA bound by specific antibodies in HEK293T cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. B, analysis for the results in (A) showing the percentage of methylated and unmethylated gene body CpG sites bound by PHF6 in HEK293T cells. The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). C, knockout (KO) of DNMT1 facilitates the occupancy of PHF6 at gene bodies. ChIP experiments were performed using primers coated on the 28S rRNA coding region in DNMT1 KO and control cells. Data are presented as the mean ± SD. D, KO of PHF6 does not alter the rDNA gene body methylation status. Genomic DNA coupled with bisulfite sequencing assays were performed to show the gene body methylation level in PHF6 KO and control cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. E, analysis for the results in (D) showing the percentage of methylated and unmethylated gene body CpG sites in PHF6 KO and control cells. The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). F, knockdown of PHF6 in DNMT1 KO and control cells by short hairpin RNAs as revealed by western blotting. TUBULIN and H3 served as a loading control. G, depletion of PHF6 partially reduces the H4K20me3 enrichment regulated by DNMT1 deficiency. ChIP experiments were performed using primers coated on the 28S rRNA coding region in the presence of shCtrl or shRNAs for PHF6 in DNMT1 KO and control cells. Data are presented as the mean ± SD. H, knockdown of PHF6 leads to increased pre-rRNA levels in DNMT1 KO cells compared with that in DNMT1 KO cells with shCtrl. RT-qPCR assays showing levels of pre-rRNA in DNMT1 KO cells with shCtrl or shRNAs for PHF6. The cycle threshold (Ct) of 45S pre-rRNA was normalized with GAPDH. Data are presented as the mean ± SD. ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed unpaired Student's t-tests.