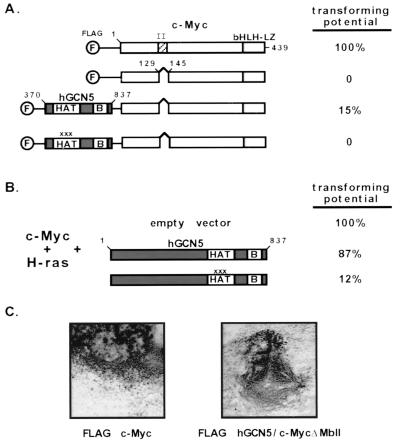
FIG. 6.
Partial rescue of a nontransforming c-Myc mutant by direct fusion to hGCN5. (A) Primary rat embryo fibroblasts were transfected with expression vectors for an activated allele of p21H-ras and one of the c-Myc constructs schematized at the left. The c-Myc constructs encoded either wild-type mouse c-Myc or a deletion mutant of this protein lacking 17 amino acids from the MbII domain. In addition, two fusion proteins were generated between the c-Myc MbII domain deletion mutant and the catalytic domain of hGCN5 (amino acids 370 to 837). For the first of these fusions, wild-type hGCN5 sequences were fused to c-Myc ΔMbII. The second fusion contained three single amino acid substitutions within the hGCN5 HAT domain (as indicated by x's). These mutations have been shown previously to block both HAT activity and transcriptional activation when introduced into the corresponding residues of S. cerevisiae GCN5. The critical bromodomain of hGCN5 (labeled B) was also included in these fusion proteins. Transforming potential of each of these proteins was determined by examining cells approximately 2 weeks posttransfection. Transforming potential was estimated based on both the total number of transformed foci generated with a given protein and the size of the individual foci obtained. bHLH-LZ, basic helix-loop-helix leucine zipper. (B) A transformation assay was performed as for panel A except that full-length versions of hGCN5 were coexpressed with c-Myc. Both wild-type and catalytically inactive forms of hGCN5 were assayed for their effect on c-Myc mediated transformation. (C) Representative foci obtained by transfection of rat embryo fibroblasts with p21H-ras and either wild-type c-Myc (left) or the MbII mutant-hGCN5 fusion protein (right).