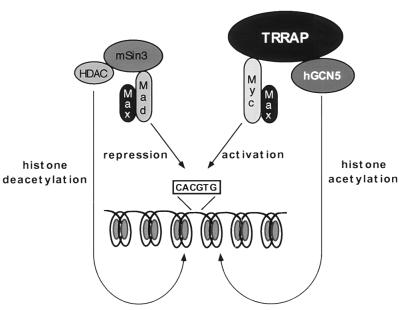
FIG. 7.
Model of the opposing biochemical functions of c-Myc and Mad. Mad family proteins dimerize with Max and repress the expression of c-myc and max target genes, thereby blocking c-Myc function. Mad-mediated repression requires the recruitment of a multiprotein complex containing histone deacetylases of the HDAC family. The demonstration here that c-Myc, through its essential cofactor TRRAP, recruits the mammalian HAT hGCN5 suggests a potential biochemical basis for antagonistic biological functions of c-Myc and Mad. In this model, c-Myc–Max heterodimers activate transcription of target genes by recruitment of TRRAP and a HAT such as hGCN5. HAT activity results in nucleosomal remodeling at target gene loci, allowing more efficient transcription. Following displacement of Myc-Max dimers from their cognate DNA recognition element, Mad-Max dimers recruit deacetylase activity to these sites, which in turn removes the acetyl groups from histones in nearby nucleosomes. This process facilitates chromatin condensation and consequently transcriptional repression.