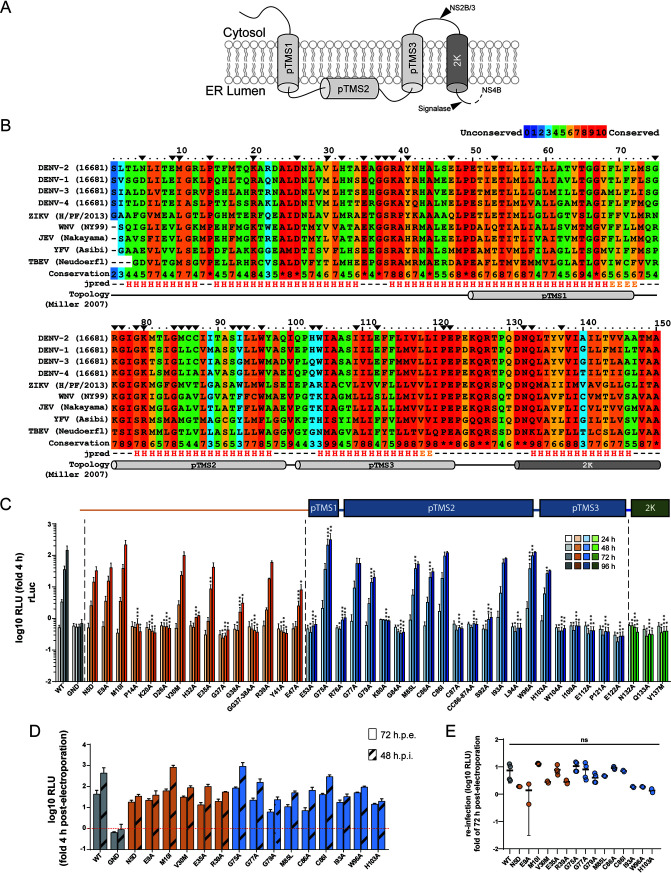
FIG 1.
Residues of DENV NS4A critical for viral replication identified by alanine-scanning mutagenesis. (A) Schematic representation of NS4A membrane topology according to reference 8. pTMS, putative transmembrane segment; 2K, 2 kDa peptide. (B) Multiple-sequence alignment of NS4A from several flaviviruses. Color grades indicate the degree of conservation according to the 0-to-10 coloring scheme given on the top right. The secondary structure prediction of each residue in context is indicated on the bottom (H, helix; E, extended). The putative transmembrane segments and 2K peptide are indicated by cylinders below the corresponding amino acid residues. Black arrowheads indicate residues selected for alanine-scanning mutagenesis. (C) Effects of mutations in NS4A on DENV replication. Mutations specified on the bottom were inserted into a DENV-2 (strain 16681) full-length reporter genome (DVs-R2A). In vitro transcripts were electroporated into Huh7 cells that were collected and lysed at different time points to measure renilla luciferase activity, which reflects RNA replication. Replication efficiency is reported as relative light units (RLU) normalized to the 4-h value (fold 4 h) that reflects transfection efficiency. Values represent the mean and SD from three independent experiments. Significance has been calculated with one-way ANOVA with Dunnett’s posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Supernatants from cells electroporated as in panel C were collected 72 h postelectroporation and used to infect Huh7 cells. Cells were lysed after 48 h, and renilla luciferase activity was measured (48 h postinfection [h.p.i.). Replication efficiency is reported as relative light units (RLU) normalized to the value determined 4 h postelectroporation. For comparison, replication levels of each construct at 72 h postelectroporation are shown (72 h.p.e.). Values represent the mean and SD from at least two independent experiments. (E) Reinfection values from panel D normalized to the respective values determined 72 h postelectroporation. Each dot represents one experiment. Significance has been calculated with one-way ANOVA with Dunnett’s posttest. ns, not significant.