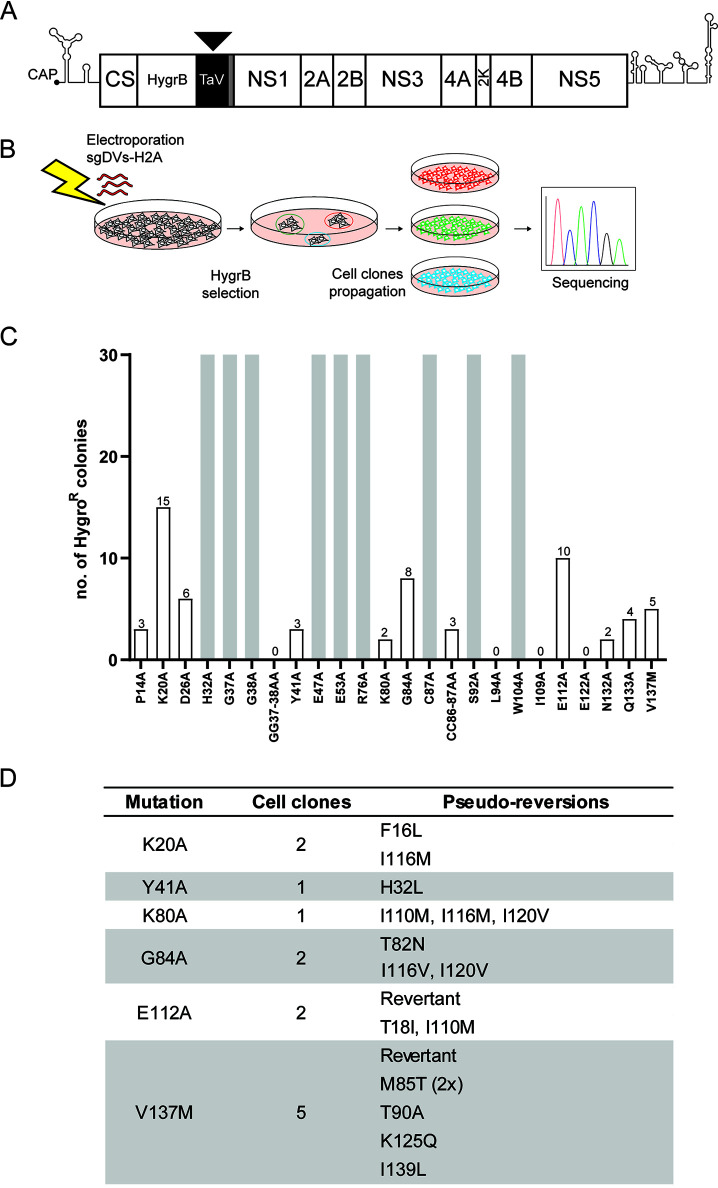
FIG 2.
Selection for pseudoreversions rescuing replication deficiency of primary mutations in NS4A. (A) Schematic representation of the selectable subgenomic DENV-2 (strain 16681) replicon (sgDVs-H2A) used to select for pseudoreversions. The hygromycin phosphotransferase gene (HygrB) is preceded by the cis-acting cyclization sequence residing in the capsid-coding region (CS). The sequence coding for the 2A-like peptide from the Thosea asigna virus (TaV) ensures cotranslational processing of the polyprotein to release properly cleaved NS1. The sequence coding for the last transmembrane helix of the envelope protein (gray box) and acting as signal sequence is retained upstream of the NS1-coding sequence to allow for ER membrane passage of NS1. (B) To select for second-site mutations rescuing viral replication, Vero E6 cells were electroporated with in vitro transcripts of sgDVs-H2A containing given primary mutations in NS4A. Electroporated cells were cultured in the presence of hygromycin B, and single-cell clones were selected and expanded. Total RNA was extracted, and coding regions of the DENV replicons were amplified by reverse transcriptase PCR (RT-PCR). Amplicons were subjected to nucleotide sequence analysis in the NS4A-coding region. (C) Number of hygromycin B-resistant cell colonies obtained after selection for each of the given constructs. Gray bars indicate confluent cell monolayers that were obtained with cells transfected with given replicon mutants. (D) List of pseudoreversions in NS4A identified in replicons after hygromycin B selection. The numbering of amino acid residues refers to strain 16681. For each cell clone, the pseudoreversion identified in replicon RNA of said clone is given. Revertant indicates a nucleotide substitution that restores the original amino acid residue.