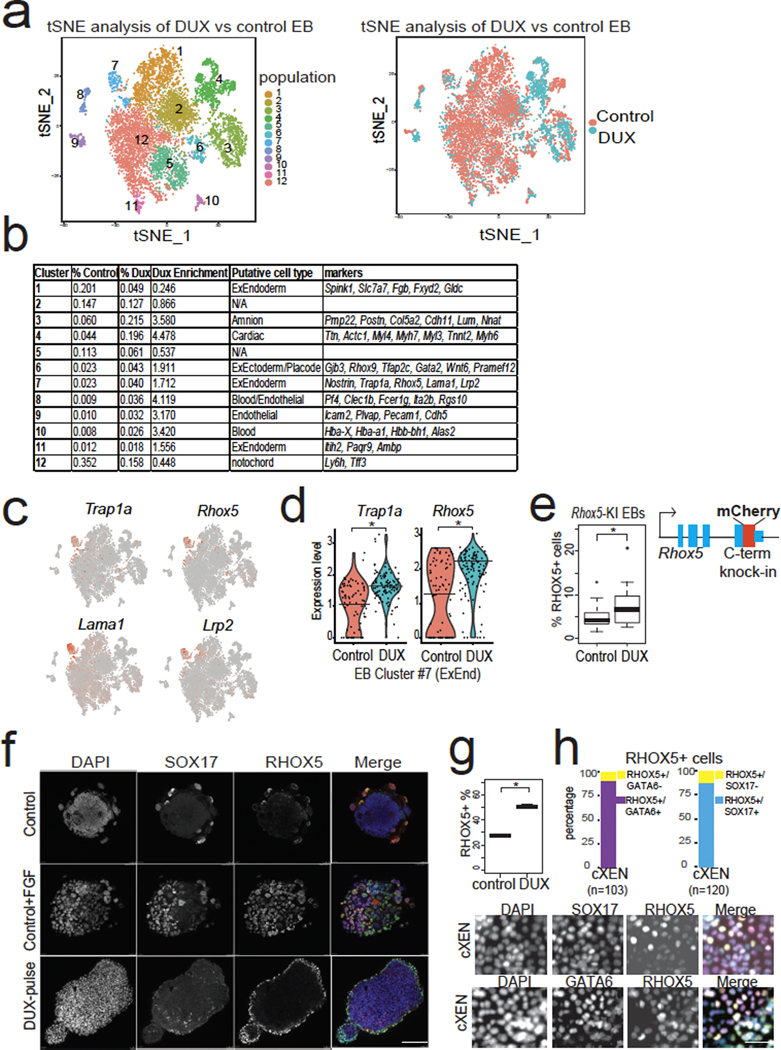
Fig. 5: Transient DUX-expression in mESCs confers increased expanded fate potential.
a, scRNA-seq identifies cell populations (clusters #1–12) from differentiated embryoid bodies (EBs) top, and scRNA-seq colored by treatment (bottom). N = 2 biological replicates per condition. tSNE = t-distributed stochastic neighbor embedding. b, Table of scRNA-seq clusters, with putative cell identity using E8.25 mouse embryo scRNA-seq34. N = 2 biological replicates per condition. c, Marker expression for cluster #7 showing canonical extraembryonic endoderm genes (ExEnd). d, Quantification of expression of Trap1a and Rhox5 (ExEnd) markers in cluster #7, showing higher levels in DUX-pulsed cells. * FDR < 0.05, Seurat. e, Quantification of % RHOX5+ cells in day 8 EBs from Rhox5-mCherry knock-in mESC clone. N = 15 biological replicates per condition, * P < 0.05, P = 0.04517, one-sided t-test. f, Immunofluorescence of EBs stained with anti-SOX17 and anti-RHOX5 antibodies. SOX17+/RHOX5+ cells can be induced in WT control EBs treated with FGF. DUX-pulsed EBs show RHOX5+ cells on the outside layer. RHOX5+ outgrowth indicated with white arrow. Representative image from n = 10 imaged EBs from each experimental condition. Merge: DAPI = cyan, RHOX5 = green, SOX17 = red. Scale bar = 150 μm. g, Extraembryonic endoderm cell (XEN) differentiation of DUX-pulsed or control mESCs, and quantification of RHOX5-mCherry-knock-in (KI) cells using flow cytometry. * P < 0.05, P = 0.001301, one-sided t-test, n = 3 biological replicates per condition. h, XEN differentiation of DUX-pulsed mESCs using Rhox5-mCherry KI clone stained with antibodies against the ExEnd marker GATA6 (n = 103 cell) or the endoderm marker SOX17 (n = 120 cells). Immunofluorescent images on bottom, with quantification of RHOX5+ cells on top. Merge: DAPI = cyan, GATA6 or SOX17 = magenta, RHOX5-mCherry = yellow. Scale bar = 50 μm. For boxplots in Figure 5e and h, the median is shown as a line in the box, and the outline of the box is depicted at the 25th and 75th percentile. The extended whiskers depict Q1 – 1.5 × IQR and Q3 + 1.5 × IQR. Outliers points are depicted as dots.