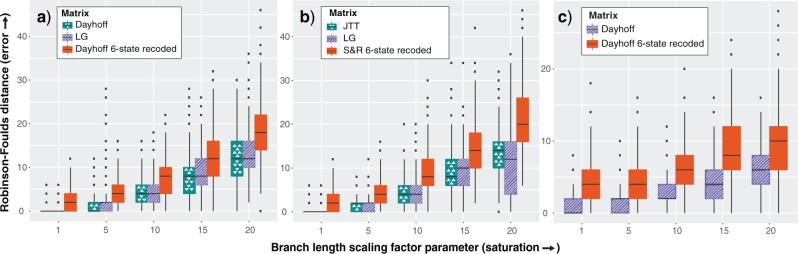
Figure 2.
Six-state recoding approaches produce more errors under increasing levels of saturation. Robinson–Foulds distances were calculated for 1000 runs for each branch length scaling factor parameter. All data were simulated on the Chang tree. a) Data sets simulated under the Dayhoff model. b) Data sets simulated under the JTT model. c) Data sets simulated under the GTR model using the amino acid rates of substitution, amino acid frequencies, and gamma rate heterogeneity estimated from the Chang data set.