Figure 4.
SETDB1 deletion caused defects in homologous pairing.
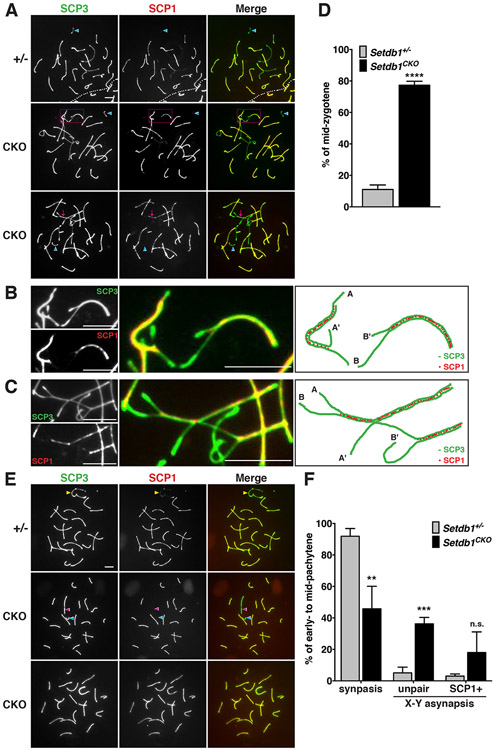
Spermatocyte spreads from Setdb1 control (+/−) and conditional knockout (CKO) testes at 16 dpp. The nuclei were stained with anti-SCP3 (green) and SCP1 (red) antibodies, indicating the axial/lateral elements and transverse filaments of the synaptonemal complex, respectively. (A) Formation of the synaptonemal complex in mid-zygotene. Y chromosome indicates by the blue arrowhead. Magenta dotted line rectangle in the middle panel indicates the asynapsed chromosomes. Magenta arrow in the bottom panel indicates the unsynapsed trivalent. (B) Higher-magnification of the middle panel in (A). The right painting illustrates asynapsed chromosomes. The A-A’ and B-B’ represent the pairing of non-homologous chromosomes that are different in length. (C) Higher-magnification of the bottom panel in (A). The right painting illustrates the unsynapsed trivalent labeled by partial synapsed A-A’ and A-B chromosomes. (D) Percentages of the abnormal mid-zygotene nuclei with synaptic defects shown in (A). (E) Formation of the synaptonemal complex in early- to mid-pachytene cells. Yellow arrowhead in the top panel indicates X and Y chromosomes synapse in the pseudoautosomal regions. Unpaired X and Y chromosomes in the middle panel indicate by the pink and blue arrowhead, respectively. The bottom panel shows both XY chromosomes or autosomes present the SCP1-positive signals. (F) Percentages of nuclei with synapsed or asynapsed X-Y chromosomes at the early- to mid-pachytene stage demonstrated in (E).
The results represent the mean ± SD of three independent experiments. (**P<0.01; ***P<0.001; ****P<0.0001; n.s., P>0.05) All scale bars represent 5 μm.