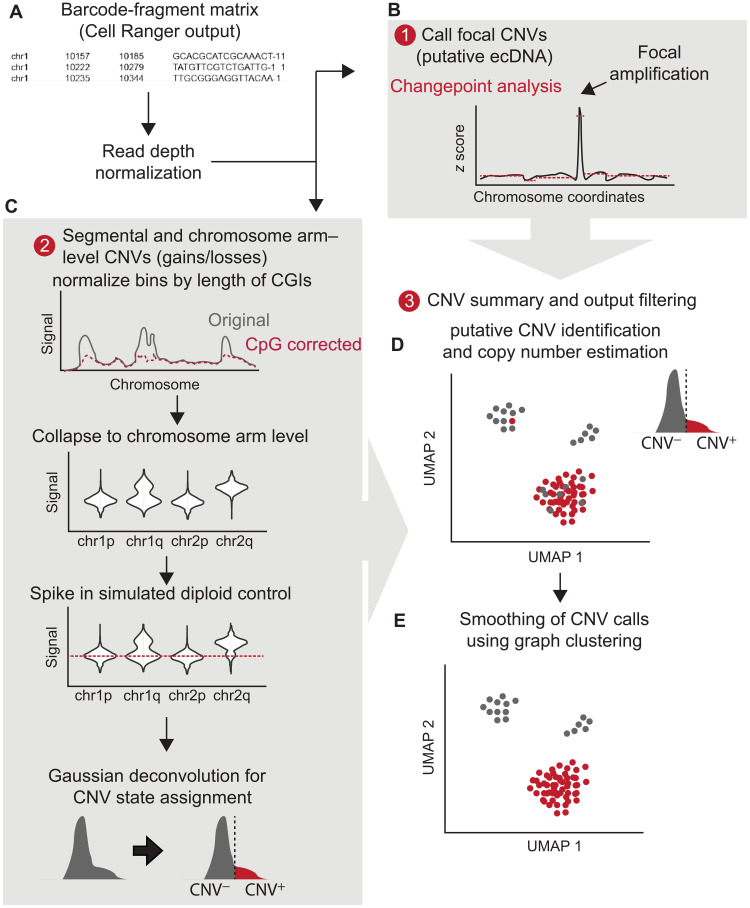
Fig. 1. Copy-scAT workflow.
(A) Copy-scAT accepts barcode-fragment matrices generated by Cell Ranger (10x Genomics) as input.(B) Large peaks in normalized coverage matrices can be used to infer focal CNVs. ecDNA, extrachromosomal DNA. (C) Normalized matrices can be used to infer segmental and chromosome arm-level CNVs. (D) Example of chromosome arm-level CNV (chromosome 10p loss) called by Copy-scAT. (E) Consensus clustering is used to finalize cell assignment.