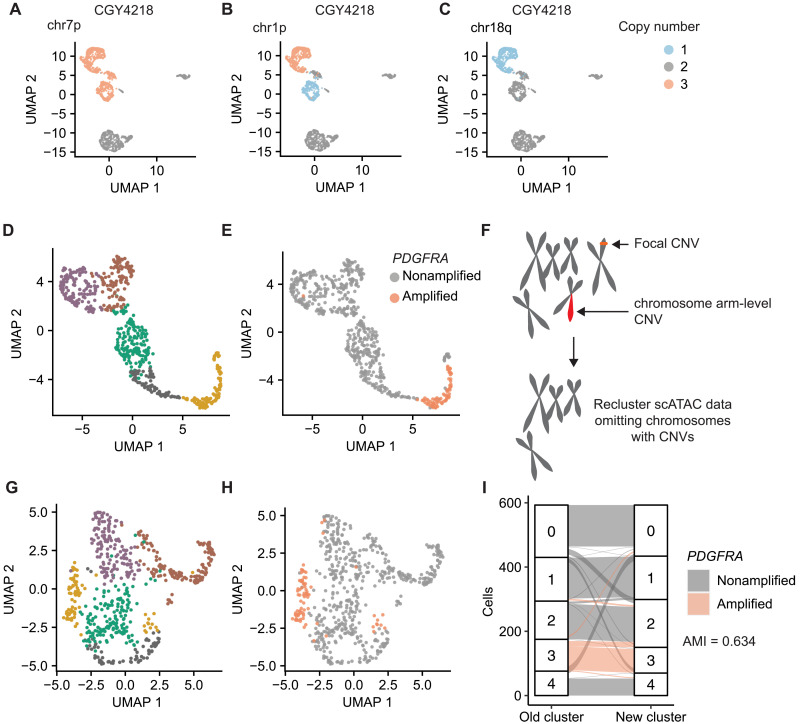
Fig. 4. Subclonal genetics influences clustering of scATAC-seq data.
(A to C) CNVs in aGBM CGY4218 segregate within specific scATAC clusters. (D and E) PDGFRA-amplified cells cluster together in aGBM CGY4349. (F) Diagram summarizing our strategy to remove CNVs from clustering of scATAC data. All chromosomes or regions with putative CNVs were removed from downstream analyses, and cells were reclustered. (G) Reclustering of (D) following removal of chromosomes and regions affected by CNVs in CGY4349. (H) Distribution of PDGFRA-amplified cells following reclustering. (I) Cluster assignments of cells in CGY4349 (aGBM specimen) before and after removal of CNV-containing regions (purple, PDGFRA-amplified cells).