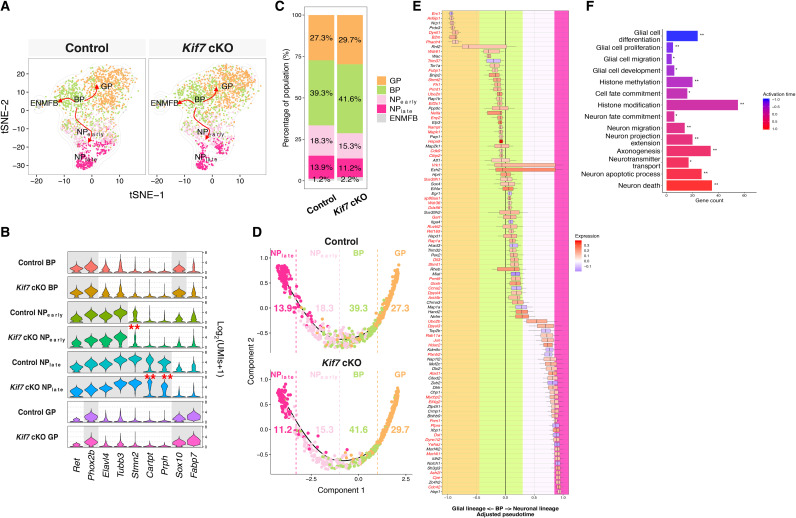
Fig. 6. Reconstruction of the differentiation trajectories of ENCCs reveals Ezh2-mediated core genes underlying neurogenic and gliogenic lineage differentiation.
(A) t-SNE projection of control and Kif7 cKO mutant cells during the ENCC differentiation process, colored by inferred cell types (GP, glial progenitor; BP, bipotent progenitor; NPearly, neuronal progenitor at early stage; NPlate, neuronal progenitor at late stage; ENMFB, enteric mesothelial fibroblast). (B) Violin plot displaying the expression of representative markers in indicated cell types. Significant expression of the genes was labeled with gray color in the background. DEGs are marked by *. (C) Bar plot shows the population structure difference between control and Kif7 cKO mutant cells. (D) Trajectory reconstruction of all single cells throughout ENCC differentiation reveals two choices of the BPs. (E) Transcriptomic timing analysis of the core gene set of 102 genes along the adjusted ENCC differentiation path. Boxes indicate the period that genes were up-regulated (red) or down-regulated (blue) in Kif7 cKO mutant cells. The background color indicated the estimated boundaries that separated the cell types. Ezh2 putative target genes were labeled in red. (F) GO analyses of the core gene set of 102 genes. The activation time of each GO term was calculated on the basis of the transcriptomic timing analysis in (E).