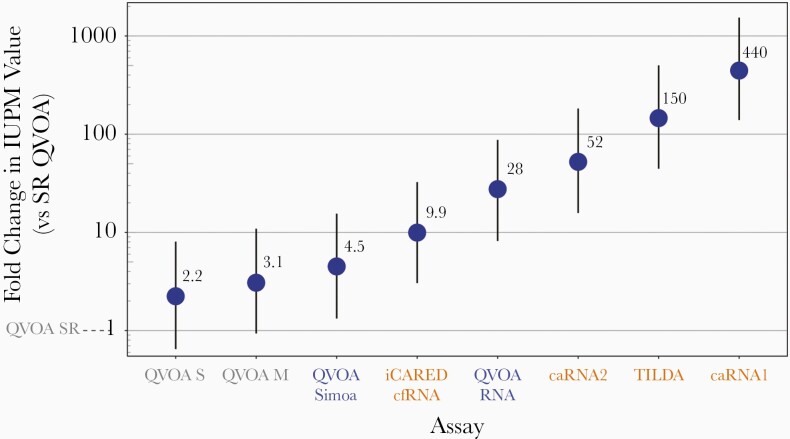
Figure 2.
Median differences with 95% credible intervals between assays presented as fold differences in scale of typical output, in infectious units per million (IUPM), for 2 classic and 6 next-generation quantitative viral outgrowth assays (QVOAs; including QVOA with ultrasensitive readout), normalized to QVOA by Southern Research (QVOA SR). The estimated fold change used the model of all 9 assays together. Classic QVOAs are shown in gray, enhanced sensitivity assays in blue, and next-generation QVOAs in orange. Abbreviations: caRNA1, cell-associated human immunodeficiency virus (HIV) gag RNA; caRNA2, cell-associated HIV tat-rev RNA; cfRNA, cell-free HIV RNA; iCARED, inducible cell-associated RNA expression in dilution; QVOA M, QVOA by University of Pittsburgh; QVOA RNA, QVOA by University of California, San Diego, with HIV RNA readout; QVOA S, QVOA by Johns Hopkins University; QVOA Simoa, QVOA by Southern Research using Simoa readout; TILDA, tat/rev-induced limiting dilution assay.