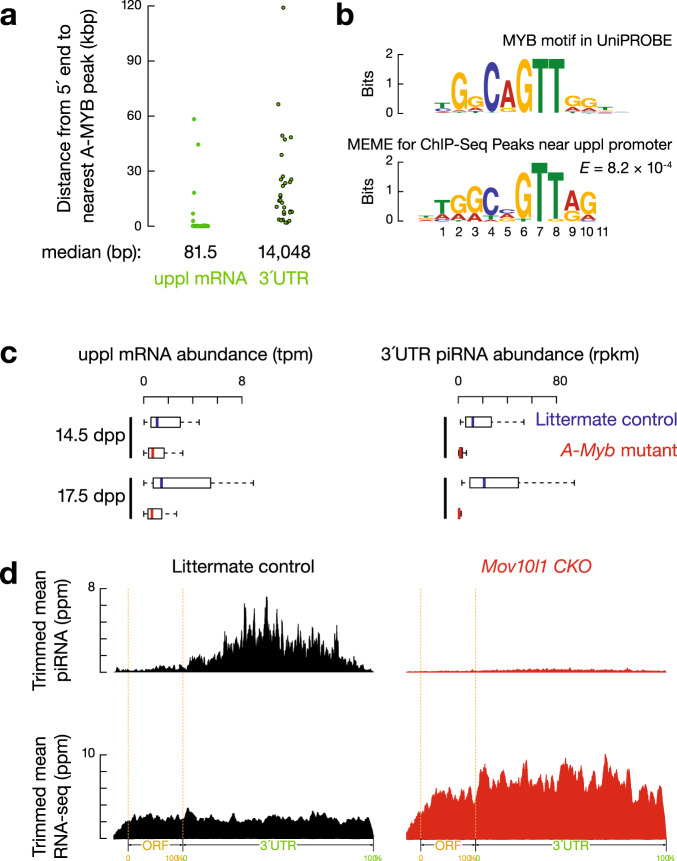
Fig. 1. mRNA precursors of 3′UTR piRNA are regulated by A-MYB.
a The distance from the annotated transcription start site of each mRNA that comes from 3′UTR piRNA-producing loci (uppl) (left, n = 30) and from the 5′-ends of uppl mRNA 3′UTRs (right, n = 30) to the nearest A-MYB ChIP-seq peak. bp: base pair, kbp: kilobase pair. b Top, the MYB motif from the mouse UniPROBE database. Bottom, MEME-identified sequence motif in the A-MYB ChIP-seq peaks near the uppl mRNA transcription state sites. E-value computed by MEME measures the statistical significance of the motif. The information content is measured in bits and, in the case of DNA sequences, ranges from 0 to 2 bits. A position in the motif at which all nucleotides occur with equal probability has an information content of 0 bits, while a position at which only a single nucleotide can occur has an information content of 2 bits. c Boxplots showing the abundance of uppl mRNA (left) and 3′UTR piRNA (right) per uppl gene in A-Myb mutants (red) and their heterozygous littermates (blue) in testes at 14.5 and 17.5 dpp. Dpp: days postpartum, tpm: transcript per million, rpkm: reads per kilobase million. uppl n = 30. Box plots show the 25th and 75th percentiles, whiskers represent the 5th and 95th percentiles, and midlines show median values. d Aggregated data for 3′UTR piRNA abundance (upper) and uppl mRNA abundance (bottom) on uppl mRNAs (10% trimmed mean) from adult Mov10l1CKO/∆ testes (left) and Mov10l1CKO/∆ Neurog3-cre (right). Signals are aligned to the transcriptional start site (5′cap) and site of polyadenylation (3′PolyA) and further aligned to the ORF regions. Dotted lines show the translation start codon (ORF Start) and stop codon (ORF End). The x-axis shows the median length of these regions. Ppm: parts per million.