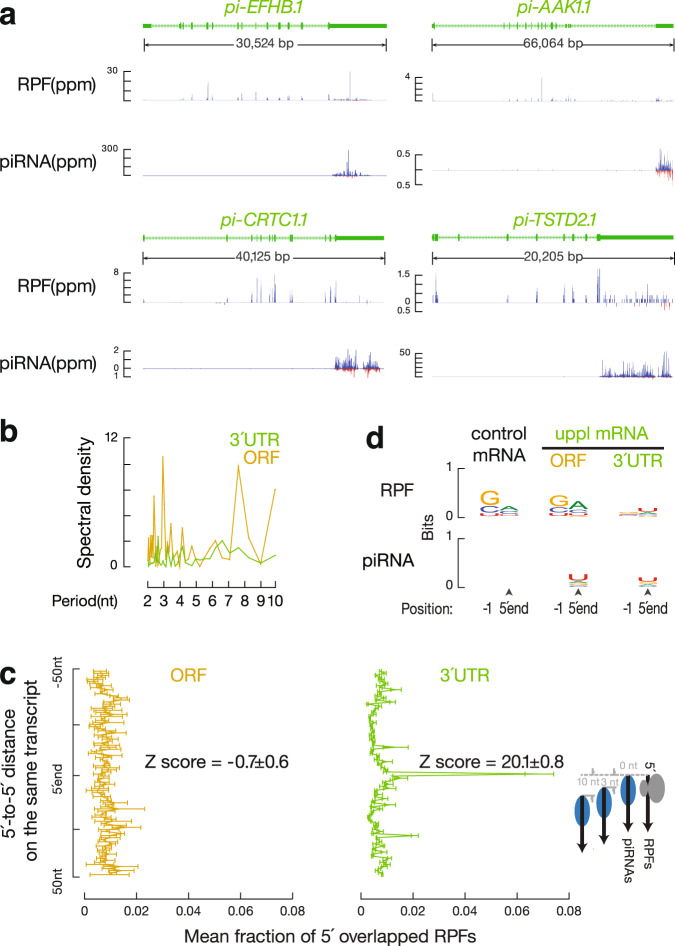
Fig. 6. Ribosome-guided 3′UTR piRNA formation is evolutionarily conserved.
a Normalized reads of RPFs and piRNAs mapping to four representative chicken uppl genes in adult rooster testes. In roosters, both strands of uppl mRNAs code for piRNAs; blue represents Watson strand mapping reads; red represents Crick strand mapping reads. b Discrete Fourier transformation of the distance spectrum of 5′-ends of RPFs across ORFs (gold) and 3′UTRs (green) of chicken uppl mRNAs in adult rooster testes. c Distance spectrum of 5′-ends of RPFs from uppl ORFs (left) and from 3′UTRs (right) that overlap piRNAs in adult rooster testes (sample size n = 3 independent biological replicates). Data are mean ± standard deviation. d Sequence logos depicting nucleotide bias at 5′-ends and 1 nt upstream of 5′-ends of the following species from adult rooster testes. Top to bottom: RPF species and piRNAs, which map to control mRNAs, uppl ORFs, and 3′UTRs, respectively.