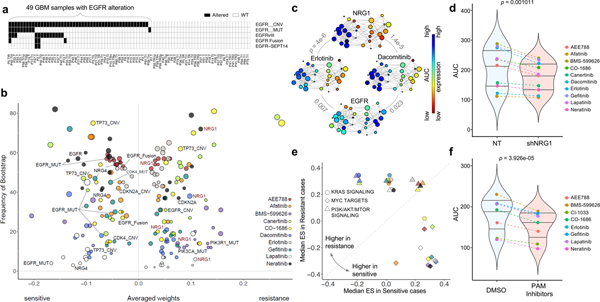
Figure 5. Predictive biomarkers for response to EGFR inhibitors in EGFR altered GBM PDCs.
(a) Mutational landscape of EGFR alterations in GBM cohort. (b) For the 10 EGFR inhibitors, top drug–feature associations identified by dNetFS are plotted for their frequency and effect size (n=49 biologically independent samples in (a). Node size is proportional to the single drug–feature linear correlation. (c) Gene expression profiles of EGFR and NRG1, and AUC drug response profiles of erlotinib and dacomitinib, over the topological representation (n=44 biologically independent samples). (d) Drug response assessment of EGFR inhibitors with shRNA-mediated knockdown of NRG1 or NT (non-target). Cell viability for each dose was normalized to shNRG1 or NT transduced cells only. (n=10 independent experiments with 3 technical replicates) (e) Comparisons of cancer pathway activities between two EGFR-altered GBM subgroups that were most sensitive and most resistance. We adopt the enrichment score derived from ssGSEA analysis as assessment. (f) Drug response assessment of EGFR inhibitors with PI3K-AKT-mTOR (PAM) inhibitors or DMSO. Cell viability for each dose was normalized to PAM or DMSO treated cells only. Mean AUC value for 4 PAM inhibitors (BYL719, BKM120, BEZ235, and AZD2014) was plotted (n=8 independent experiments with 3 technical replicates). P-values: c, pearson correlation test. P-values: d,f, two-sided wilcox rank sum test. Horizontal lines within the violin plot represent 0.25, 0.50, and 0.75 quantiles.