Figure 3.
Comparison of iINs, mINs, and orgINs
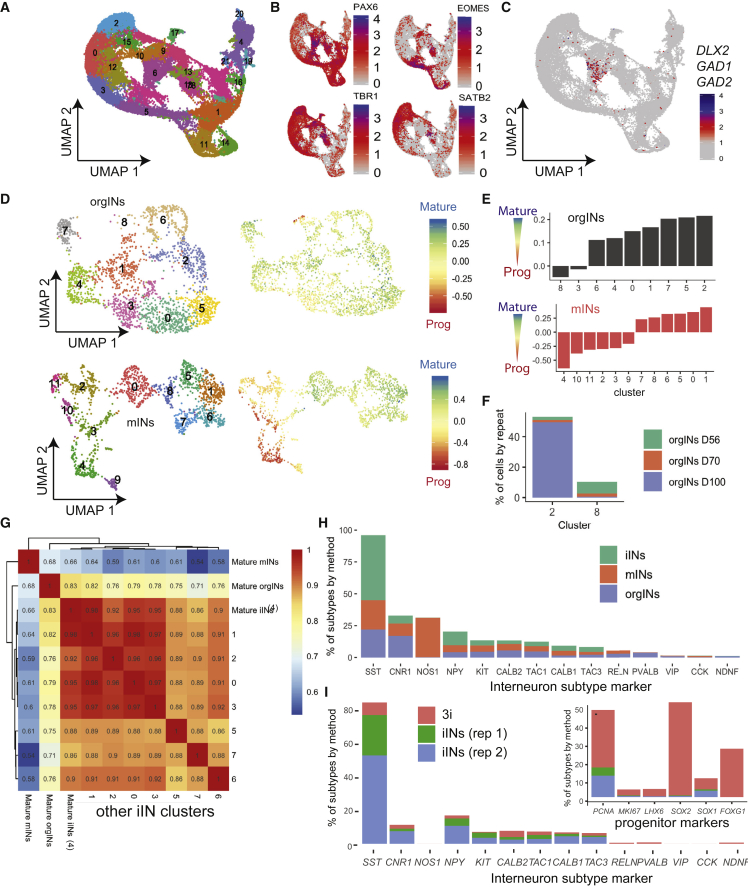
(A) Unsupervised UMAP clustering of 36,076 single cells derived from day 56 (D56) unfused, day 70 (D70) and day 100 (D100) fused cortex and MGE organoids.
(B) UMAP from (A) colored by the normalized expression of indicated genes in individual cells.
(C) As in (B), except that the plot shows the co-expression of indicated interneuron marker genes, indicating that cluster 6 is expressing these markers together.
(D) (Top) Cells from interneuron cluster 6 of the organoid scRNA-seq data in (A) (orgINs) were divided into nine interneuron clusters (0–8) and plotted on a UMAP (left). In the same UMAP to the right, individual cells are colored by the maturation score from Close et al. (2017). (Bottom) UMAP of the scRNA-seq data from the mIN dataset of 2D cultures of interneurons made by directed differentiation (Close et al., 2017), separating interneurons into 12 distinct clusters (0–11) (left) and colored by the maturation score from (Close et al., 2017) (right).
(E) Maturation score for the clusters in (D), ordered from least to most mature.
(F) The contribution of cells from different organoid harvest time points to the orgINs clusters with highest (cluster 2) and lowest (cluster 8) maturation state.
(G) Pearson correlation analysis for the most mature orgIN cluster (cluster 2 from (E)), the most mature mIN cluster (cluster 1 from (E)), and all iIN clusters (AD-method-derived interneurons) from Figures 1 and 2 (with cluster 4 containing the most mature cells).
(H) A survey of the subtypes of interneurons found in iINs, mINs, and orgINs.
(I) A similar analysis as in (H), but for individual experiments for directed differentiation (3i) of human induced PSCs or the two biological replicates of iINs from hESCs.