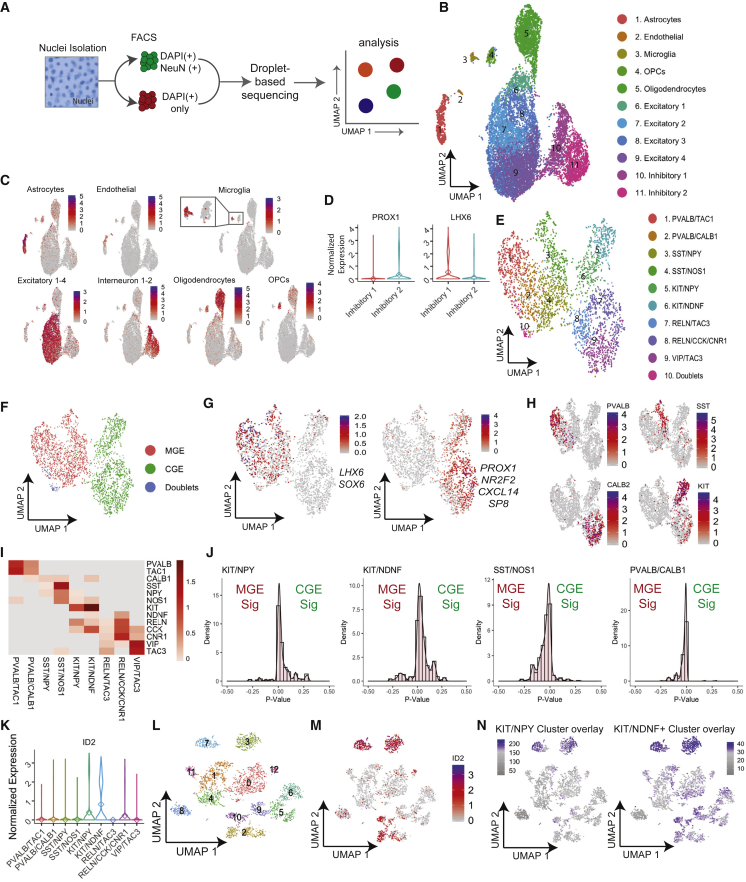
Figure 4.
Profiling interneurons from human adult brains with snRNA-seq
(A) Schematic of the workflow for snRNA-seq from frozen archived adult brain cortex.
(B) UMAP of 17,879 nuclei from six adult cortexes. Cell identities were assigned to clusters based on marker gene expression.
(C) Simultaneous expression of several genes known to define clusters in (B).
(D) Violin plots for the normalized expression of PROX1 and LHX6 in the two interneuron clusters 10 and 11 in (B), called Inhibitory 1 and 2, to decipher MGE- versus CGE-derived interneurons.
(E)The UMAP of the cells from interneurons from clusters 10/11 in (B), revealing 10 interneuron sub-clusters (1–10) of the adult brain, labeled based on key marker expression.
(F) UMAP from (E) split into MGE- versus CGE-derived interneurons based on simultaneous expression of region-specific markers.
(G) UMAP as in (E) showing the simultaneous expression of established markers of either MGE (LHX6, SOX6) or CGE (PROX1, NRF2, CXCL14, SP8).
(H) Normalized expression of indicated interneuron subtype marker on the UMAP from (E).
(I) Heatmap of normalized expression of selected interneuron markers by clusters from (E).
(J) Histograms to demonstrate the developmental origin of indicated KIT+ clusters and SST/NOS1, PVALB/CALB1 clusters as controls. MGE and CGE gene signatures were generated by taking the top 20 most differentially expressed genes between MGE- and CGE-derived interneurons. Individual cells were assigned MGE and CGE scores based on their individual expression of each MGE or CGE gene, using the Wilcoxon test.
(K) Normalized expression for ID2 in interneuron sub-clusters from in (E).
(L) UMAP of all interneuron subtypes in a human brain dataset from the Allen Brain Database.
(M) As in (L), with an overlay of the normalized expression of ID2.
(N) Simultaneous expression of KIT/NPY and KIT/NDNF cluster signature genes (genes unique to KIT/NPY or KIT/NDNF clusters from E with an average log2 fold-change cutoff of 0.5), overlayed on the UMAP in (L).