Abstract
MRI-derived features of presumed cerebral small vessel disease are frequently found in Alzheimer’s disease. Influences of such markers on disease-progression measures are poorly understood. We measured markers of presumed small vessel disease (white matter hyperintensity volumes; cerebral microbleeds) on baseline images of newly enrolled individuals in the Alzheimer’s Disease Neuroimaging Initiative cohort (GO and 2) and used linear mixed models to relate these to subsequent atrophy and neuropsychological score change. We also assessed heterogeneity in white matter hyperintensity positioning within biomarker abnormality sequences, driven by the data, using the Subtype and Stage Inference algorithm. This study recruited both sexes and included: controls: [n = 159, mean(SD) age = 74(6) years]; early and late mild cognitive impairment [ns = 265 and 139, respectively, mean(SD) ages =71(7) and 72(8) years, respectively]; Alzheimer’s disease [n = 103, mean(SD) age = 75(8)] and significant memory concern [n = 72, mean(SD) age = 72(6) years]. Baseline demographic and vascular risk-factor data, and longitudinal cognitive scores (Mini-Mental State Examination; logical memory; and Trails A and B) were collected. Whole-brain and hippocampal volume change metrics were calculated. White matter hyperintensity volumes were associated with greater whole-brain and hippocampal volume changes independently of cerebral microbleeds (a doubling of baseline white matter hyperintensity was associated with an increase in atrophy rate of 0.3 ml/year for brain and 0.013 ml/year for hippocampus). Cerebral microbleeds were found in 15% of individuals and the presence of a microbleed, as opposed to none, was associated with increases in atrophy rate of 1.4 ml/year for whole brain and 0.021 ml/year for hippocampus. White matter hyperintensities were predictive of greater decline in all neuropsychological scores, while cerebral microbleeds were predictive of decline in logical memory (immediate recall) and Mini-Mental State Examination scores. We identified distinct groups with specific sequences of biomarker abnormality using continuous baseline measures and brain volume change. Four clusters were found; Group 1 showed early Alzheimer’s pathology; Group 2 showed early neurodegeneration; Group 3 had early mixed Alzheimer’s and cerebrovascular pathology; Group 4 had early neuropsychological score abnormalities. White matter hyperintensity volumes becoming abnormal was a late event for Groups 1 and 4 and an early event for 2 and 3. In summary, white matter hyperintensities and microbleeds were independently associated with progressive neurodegeneration (brain atrophy rates) and cognitive decline (change in neuropsychological scores). Mechanisms involving white matter hyperintensities and progression and microbleeds and progression may be partially separate. Distinct sequences of biomarker progression were found. White matter hyperintensity development was an early event in two sequences.
Keywords: Alzheimer’s, biomarkers, cerebrovascular disease, microbleeds, white matter hyperintensities
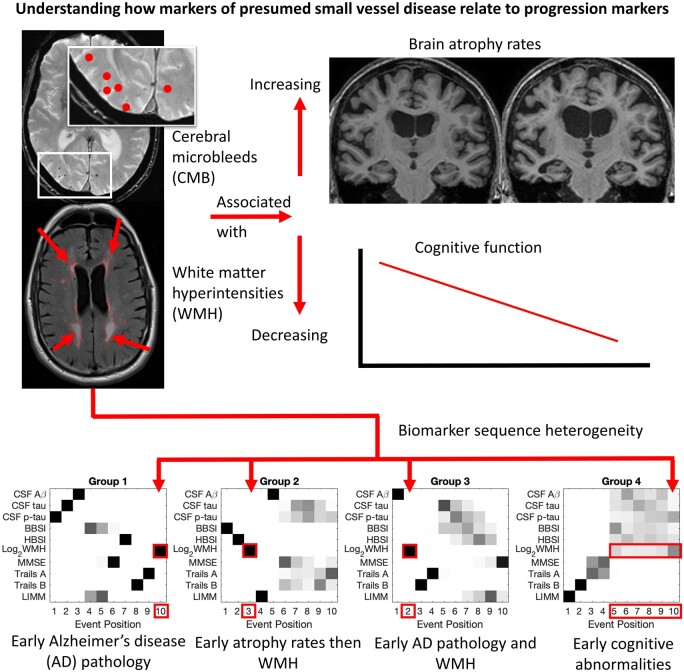
Fiford et al. report that presumed small vessel disease markers (white matter hyperintensities and cerebral microbleeds) independently predict increased brain atrophy and cognitive decline. Data-driven analysis of sequences of biomarker abnormality identified distinct groups, two of which had white matter hyperintensity development as an early event.
Graphical Abstract
Graphical Abstract.
Introduction
White matter hyperintensities (WMHs), lacunes and cerebral microbleeds (CMBs) are features of presumed cerebral small vessel disease (SVD) which increase with age and are often found in Alzheimer’s disease.1–4 Different SVD markers likely reflect different disease processes and vascular pathologies and may independently contribute to neurodegeneration and cognitive decline. This study investigates whether SVD markers are associated with Alzheimer’s disease progression measures in a cohort including normal ageing and all putative stages of Alzheimer’s disease. The study also assesses whether SVD is an early marker of disease relative to classical Alzheimer’s disease measures. Together, this allows for a better understanding of the presumed vascular contributions to Alzheimer’s disease.
Disease progression is often assessed using measures of cognitive decline and neurodegeneration. Cognitive decline is measured using changes in neuropsychological scores, whereas a commonly used proxy of neurodegeneration is brain atrophy rates measured using serial MRI.5 WMHs are associated with higher atrophy rates in normal ageing and mild cognitive impairment (MCI).6–8 Contrastingly, the association of CMBs with atrophy rates is poorly understood; one study found no association between brain volume change prior to CMB development.9
Data-driven techniques give insight into another feature of progression: the sequence in which biomarkers become abnormal.10 These techniques can provide information regarding the heterogeneity and uncertainty of different biomarker progression sequences and possible sub-group aetiologies. This information is crucial in understanding whether multiple disease-progression models, which include presumed vascular markers, are needed.
This study investigated SVD markers in a cohort including normally ageing individuals, those with significant memory concern (SMC), early and late cognitive impairment (EMCI and LMCI) and clinical Alzheimer’s disease. We assessed independent relationships of baseline WMHs and CMBs with: (i) brain and hippocampal atrophy rates using the boundary shift integral (BSI); and (ii) changes in neuropsychological scores. We also performed analyses to determine clusters of individuals that follow different temporal sequences of biomarkers becoming abnormal. We hypothesized that SVD measures would independently and partially predict progression measures, and that a data-driven approach would reveal heterogeneity in biomarker progression sequences.
Methods
Subjects
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). ADNI was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early Alzheimer’s disease. For up-to-date information, see www.adni-info.org Accessed 07 October 2021. Newly enrolled clinically defined participants from ADNI2 and ADNIGo (ADNI2/GO) were analysed in this study. This included controls, SMC, EMCI, LMCI and Alzheimer’s disease. SMC were included since they are at higher risk of decline and may be different to controls without memory concerns.
Inclusion criteria for all individuals were that they were good general health, were between 55 and 90 (inclusive) years of age, had a reliable study partner, could speak English or Spanish, and had Hachinski score < 5.11 Controls, SMC, EMCI and LMCI had to have Mini-Mental State Examination (MMSE) scores were between 24 and 30 (inclusive). Controls and SMC had to have a clinical dementia rating (CDR) of 0, both MCI groups had to have a CDR of 0.5. Controls, SMC, EMCI and LMCI had to have preserved activities of daily living and an absence of dementia. SMC, EMCI, LMCI and Alzheimer’s disease had to have subjective memory concerns. Controls and SMC had to be normally functioning as measured by education-adjusted scores on delayed recall of one paragraph from Wechsler Memory Scale Logical Memory II.
Specific inclusion criteria were in place for all groups. Controls had to have no memory complaints. SMC had to have a cognitive change index score of more than 15. EMCI subjects were separated from LMCI subjects on the Wechsler Memory Scale Logical Memory II. EMCI had to have scores for ≥16 years of education: 9–11; for 8–15 years of education: 5–9; or for 0–7 years of education: 3–6. LMCI had to have scores for ≥16 years of education of ≤8; for 8–15 years of education: ≤4; for 0–7 years of education: ≤2. Alzheimer’s disease participants had MMSE between 20 and 26 inclusive, a CDR of 0.5 or 1.0 and had to meet NINCDS/ADRDA criteria for probable Alzheimer’s disease.
For inclusion in this study, individuals required good quality serial T1-weighted MRI necessary for atrophy rate measurement, a good quality SPM12 (https://www.fil.ion.ucl.ac.uk/spm/ Accessed 07 October 2021) segmentation to generate total intracranial volume (TIV) as well as a good quality baseline WMH measurement.
Imaging
All images were from 3 T scanners and underwent standard quality control at the Mayo Clinic (Rochester, MN, USA), which included protocol compliance check, inspection for clinically significant structural abnormalities, and image quality assessment. This study utilized unprocessed baseline non-accelerated T1-weighted MRI and baseline FLAIR and T2* images (for SVD measurement). For estimation of atrophy rates serial accelerated T1-weighted MRI imaging12 was used which had preprocessing performed at Mayo including correction of gradient warping13 and reduction of image inhomogeneity.14 Individual accelerated T1-weighted images were additionally checked at the Dementia Research Centre (DRC), UCL, London, UK, for significant motion artefacts that would cause blurring at tissue boundaries. T1-weighted images were near-isotropic, FLAIR images had a slice thickness of 5 mm, and T2* of 4 mm (see http://adni.loni.usc.edu/methods/documents/mri-protocols/ Accessed 07 October 2021). For atrophy rate estimation, only images obtained using the same scanner as the baseline assessment were used.
SVD marker detection
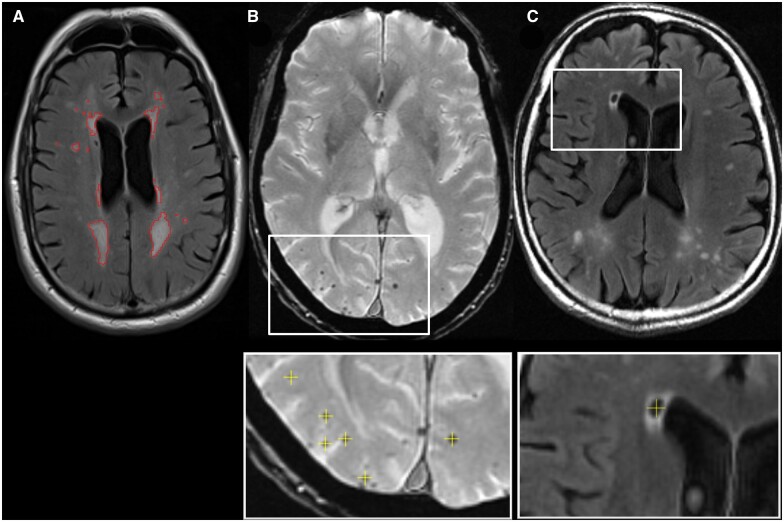
SVD measurement examples are shown in Fig. 1. Supratentorial white matter and deep grey matter hyperintensities (WMH) were estimated on FLAIR images, in combination with the T1-weighted image, using BaMoS.15 This automated technique segments WMHs by modelling each tissue class (grey, white, CSF and non-brain) as a mixture of Gaussians whose number is automatically and dynamically determined using a split-and-merge strategy and constrained by anatomical probabilistic atlases and neighbourhood constraints (using Markov Random Fields). Both a skull-stripping mask and subject-specific statistical tissue priors were obtained as a result of the label-fusion GIF framework.16 After convergence of the models, candidate lesion voxels from the outlier components of the data model were selected based on their location and intensity compared with healthy-appearing white matter. Connected components of these voxels were then automatically classified as lesion or artefact based on anatomical and shape rules. The final WMH volume is obtained from the integration of the probability maps of selected lesions, with the probabilities defined based on the level of outlierness compared with healthy white matter. All WMH binary masks were inspected by an experienced rater.
Figure 1.
Small vessel disease markers. Axial views of A white matter hyperintensity (outlined in red) on a FLAIR image; B Cerebral microbleeds unlabelled with labelling shown underneath (yellow crosses) on a T2* image; C a lacune with labelling shown underneath (yellow cross) on a FLAIR image.
For CMB labelling, FLAIR and T1-weighted images were linearly registered to the T2* space using NiftyReg17 to ensure that the lower-resolution T2* remained in native space. CMBs were identified on T2* images with co-registered FLAIR and T1-weighted images available for reference using the open-source viewer NiftyMidas.18 During identification, CMBs were counted and marked (see Fig. 1). To be considered a CMB, the hypointensity on T2* had to be small, <10 mm in diameter, well-defined, and either ovoid or round. The CMB had to be blooming on the T2* compared with the other imaging sequences. CMB mimics, including vessels, mineralization, partial volume and air-bone interface artefacts,19 were identified using all imaging types, and disregarded. Care was taken to ensure the same CMB was not counted on multiple slices. One rater marked all CMBs, with the manual counts performed according to the MARS scale.19 Hypointensities on T2* that were difficult to confirm as CMBs were checked with a neuroradiologist.
Lacunes of presumed vascular origin4 were identified on FLAIR images in a similar manner to CMBs. Co-registered T1-weighted to FLAIR images were used to aid lacune identification in a similar process to CMBs. A lacune was recorded and marked using NiftyMidas (see Fig. 1) if a hypointense area on the FLAIR images corresponded to a region of hypointensity on the registered T1-weighted images with CSF-like signal on both. These often had a hyperintense rim on FLAIR. Lacunes were only included in the regions of white matter in the territory of perforating arterioles: specifically those from the posterior cerebral artery and the middle cerebral artery. The size of the lacune was important4; there was a lower limit of ≥3 mm and an upper limit of ≤15 mm in diameter as inclusion criteria. All lacunes were checked by a neuroradiologist.
Whole-brain segmentations were generated using Brain MAPS20 and hippocampal regions using STEPS.21 Regions were checked and edited where necessary by experienced raters at the DRC. Symmetric whole-brain and hippocampi BSIs were calculated following registration into a within-subject group-wise space.22 Changes in volume from baseline were used in analyses. TIV was calculated using a published method which sums the tissue classes of CSF, grey matter and white matter using SPM12.23
Neuropsychological, demographic and CSF variables
MMSE, Trails A and B, and logical memory—immediate recall (LIMM) were downloaded. These tests cover general cognition with a memory bias (MMSE), processing speed (Trails A), executive function (Trails B) and logical memory (LIMM). Baseline age, APOE ε4 status, history of hypertension and smoking, height and weight, diabetes status, years in education, gender and CSF amyloid beta (Aβ 1–42), total tau (tau) and phosphorylated (ptau) were downloaded. CSF measures were those from the micro-bead-based multiplex immunoassay, the INNO-BIA AlzBio3 RUO test (Fujirebio, Ghent, Belgium) on the Luminex platform. Baseline geriatric depression scale total scores and change in diagnosis identifications for the 12-month time-point were downloaded.
Statistical analysis
Variables of interest
Outcomes of interest modelled separately were (i) neurodegeneration measures: brain and hippocampal atrophy rates; and (ii) cognitive decline measures: changes in LIMM, MMSE, Trails A and Trails B. Predictors of interest were baseline: WMH volumes and presence of CMBs. These predictors were used both in separate models, and, also together to assess independence of prediction of outcomes.
Variable transformation
WMHs were log2 transformed, owing to skewness. Body mass index (BMI) was calculated from height and weight variables (kg/m2). CMBs were treated as binary variables, indicating the presence of at least one CMB. For CMBs, the count data were not normally distributed with the majority of individuals having either no CMBs or a single CMB and therefore we chose to investigate CMB as a binary variable. The inverse of Trails A and Trails B was used in the linear mixed-effects models described below to improve model fit.
Analysis of baseline demographic, neuropsychological and imaging variables
STATA v13 or later was used for all statistical tests. All tests were two-tailed and the alpha-level used was P < 0.05. P-values in this first set of analyses represent differences between diagnostic groups. Fisher’s exact test was used to assess differences in distribution of gender and APOEε4 carrier (binary) status, presence of lacunes and CMB, diabetes status, and history of hypertension across diagnostic groups. Differences according to smoking and race were examined using chi2 tests. For all other variables, linear regression was used to compare the diagnostic groups at baseline adjusting for relevant confounders by including them as additional predictors in the model. Comparison of log2WMH was adjusted for TIV and comparison of brain volume, and hippocampal volume was adjusted for age, gender and TIV as these have been found to be confounders in cross-sectional analyses.24 Comparison of neuropsychological scores was adjusted for age, gender and education as these are known cross-sectional confounders.25
Atrophy rates with SVD predictors
For brain and hippocampal volume change, linear mixed-effects models for directly measured change were fitted on the whole dataset.26 Brain or hippocampal atrophy rate, was the outcome, with WMH or CMB variables as predictors. Atrophy was measured using available T1-weighted MRI, from screening to 3-, 6-, 12-, 24-month interval scans, then annual assessments beyond 24-months for all groups except the Alzheimer’s disease group. The mixed-effects models allow for missing atrophy rate measures. Interactions between WMH or CMB markers and time were used to estimate associations between SVD and rate of atrophy (the outcome of interest). Separate models for each type of atrophy were fitted, as well as for WMH or CMBs as predictors. The effects of lacunes were not reported owing to low prevalence. Models for each outcome including all three markers of SVD were also fitted. All models were adjusted for TIV and diagnostic group. To account for repeated measures, random slopes and intercepts at the level of the individual were included. A different random slope and intercept variance was estimated for each diagnostic group, to allow for different levels of variation in atrophy and atrophy rate between groups. We further fitted interactions between WMH or CMB, time and diagnostic group to test for the differential effect of either WMH or CMB measurements on atrophy rates between diagnostic groups. We fitted another pair of models including both CMB and WMH allowing an interaction between the main effects of CMB and WMH and the effect of CMB and WMH on the rate of change in the outcomes to assess for a multiplicative effect of the presence of CMBs on the effect of WMH on the outcome of interest.
Psychology test modelling with SVD predictors
Linear mixed-effects models were used to assess whether the WMH and CMB markers predicted changes in neuropsychological scores. Scores were the outcome, with WMH and CMB variables as predictors. Scores were used from screening or baseline, 6-, 12-month intervals then annual assessments for all tests apart from LIMM which was not assessed at 6-months. Individuals with Alzheimer’s disease were not followed beyond 24 months. The mixed-effects models allow for missing neuropsychological scores. Interactions between WMH and CMB markers and time were used to estimate associations between these SVD markers and neuropsychological score change (the outcome of interest). Separate models for each score were fitted, as well as for WMH and CMB separately. Models for each outcome including all three markers of SVD were also fitted. All models included random slopes and intercepts at the level of the individual. Different random effect variances were estimated for control, Alzheimer’s disease and pooled MCI and SMC groups. Correlations between intercepts and slopes were permitted by using unstructured covariance matrices within each group. Where models did not converge the random effects structure was simplified by combining the control, MCI and SMC groups (see Supplementary methods). For MMSE, inference was based on bias-corrected and accelerated bootstrap confidence intervals from 2000 replications since it was not possible to transform MMSE to allow the residuals to follow a normal distribution. We fitted further models to assess evidence for the differential effect of WMH and CMBs on score change between diagnostic groups. These models included as predictors: WMH or CMB; interaction between WMH or CMB measure and diagnostic group; and a three-way interaction between the WMH or CMB, diagnostic group and time. We fitted another set of models including both CMB and WMH allowing an interaction between the main effects of CMB and WMH and the effect of CMB and WMH on the slopes of the outcomes to assess whether there was a multiplicative effect of CMB presence on the effect of WMH on neuropsychological scores.
Disease-progression measurement modelling
Individuals with complete data for all baseline measures of interest and baseline to 12-month atrophy rate and neuropsychological tests were used. The Subtype and Stage Inference (SuStaIn) algorithm27 was used to determine groups of individuals with distinct progression sequences. Progression sequences are orderings of biomarkers becoming abnormal, i.e. an event-based model (EBM).10,28 An important feature of this approach is that the sequence of events is estimated from the data, rather than relying on a priori clinical staging. Furthermore, the uncertainty of sequences can be ascertained. The occurrence of each event is informed by a probability measure, estimated by fitting a mixture of two Gaussians to each biomarker to determine the likelihood a biomarker measurement is ‘normal’ or ‘abnormal’. The SuStaIn algorithm is then used to simultaneously cluster individuals into groups, estimate an event sequence for each group, and derive a biomarker severity stage (SuStaIn stage) for each subject. The uncertainty in the sequence of the events for each cluster is then determined using a Markov Chain Monte Carlo (MCMC) algorithm. This produces a single most likely sequence as well as a relative likelihood of other sequences enabling assessment of uncertainty.28
Only single continuous measurements for a variable of interest can be used in this approach, such as baseline measures and change metrics (difference between two time-points). CSFAβ1–42, tau and ptau, whole-brain BSI, hippocampal BSI, log2WMH and first assessment MMSE, Trails A, Trails B and LIMM were used. Measures such as presence of CMBs, and changes in neuropsychology scores were not included due to the binary nature of the CMBs and variance in neuropsychological score change. Between-cluster differences were then assessed for variables of interest not included in the models. We assessed differences in the initial diagnostic group, SuStaIn stage, baseline age, gender, years of education, hypertension, smoking history, BMI, APOE ε4 carrier status, presence of CMBs, brain volume, hippocampal volume, geriatric depression score, and annualized changes calculated from the baseline and 12-month values of MMSE, Trails A, Trials B, LIMM. The tests used included chi2 for diagnostic group at baseline, SuStaIn stage and smoking status. For gender, history of hypertension, APOE ε4 status, presence of microbleeds and diabetes status, Fisher’s exact test was used. For age, education, BMI, neuropsychological change scores, brain volumes, hippocampal volumes and geriatric depression scale total scores, linear regression was used. TIV, age and gender were used to adjust the analysis of brain and hippocampal volumes across clusters. Gender was used to adjust the analysis of years in education. We also report the percentage of individuals who change diagnostic group between baseline and the 12-month time-point.
To aid interpretability we also report mean (SD) variables entered into SuStaIn EBM by subtype (derived group) without statistical testing across grouping.
Data availability
ADNI data are available for download for approved researchers (www.adni-info.org Accessed 07 October 2021). The NiftyMidas visualization and labelling software is available (https://github.com/NifTK/NifTK Accessed 07 October 2021). NiftyReg (used for the linear registration) is available (https://github.com/KCL-BMEIS/niftyreg/wiki Accessed 07 October 2021).
Results
Subjects
Seven hundred and thirty-eight subjects with good-quality WMHs, TIVs and brain atrophy rate measurements were included in the main analysis, see Fig. 2. A subset of those with WMH segmentations had complete lacune (n = 731) and CMB data (n = 717). For hippocampi, a subset of individuals with hippocampal volumes changes were available with WMH data (n = 717), lacune data (n = 710) and CMB data (n = 697).
Figure 2.

Flowchart depicting subject selection for analysis. Definitions: white matter hyperintensity (WMH); statistical parametric mapping (SPM); total intracranial volume (TIV).
Demographics
Participant groups differed in age, with the EMCI as the youngest (71.4 years) and Alzheimer’s disease (75.0 years) as the oldest group (see Table 1). The Alzheimer’s disease group also had the highest frequency of APOE ε4 carriers, the lowest level of education and the lowest BMI. There was no evidence of any differences in race, diabetes or gender proportions between groups.
Table 1.
Subject demographics and basic imaging information for the ADNI2/Go cohort
| C | SMC | EMCI | LMCI | Alzheimer’s disease | Group difference (P-value) | |
|---|---|---|---|---|---|---|
| N | 159 | 72 | 265 | 139 | 103 | |
| Age at baseline, years | 73.7 (6.2) | 72.0 (5.6) | 71.4 (7.3) | 71.9 (7.7) | 75.0 (7.8) | <0.001 |
| Male (%) | 46.5 | 44.4 | 54.3 | 54.0 | 59.2 | 0.2 |
| Hypertension (% hypertensive) | 51.6 | 41.7 | 50.9 | 44.6 | 45.6 | 0.4 |
| Smoking (% never/historical/current) | 60.4/35.9/3.8 | 52.8/44.4/2.8 | 60.4/38.1/1.5 | 67.6/30.2/2.2 | 64.1/32.0/3.9 | 0.4 |
| Body mass index, kg/m2 | 27.2 (4.3)b | 28.0 (6.4)a | 28.0 (5.3) | 27.4 (5.0) | 25.6 (4.2) | 0.001 |
| Percentage with diabetes | 6.9 | 12.5 | 11.3 | 8.6 | 8.8a | 0.5 |
| Percentage APOE ε4 carriers | 28.9 | 37.5 | 42.3 | 56.8 | 73.8 | <0.001 |
| Years of education | 16.5 (2.5) | 17.0 (2.4) | 16.0 (2.7) | 16.7 (2.5) | 15.7 (2.7) | <0.001 |
| First assessment MMSE | 29.0 (1.3) | 29.0 (1.2) | 28.3 (1.6) | 27.6 (1.9) | 23.1 (2.1) | Not appropriate |
| First assessment Trails A | 33.6 (10.5) | 34.7 (12.7) | 36.0 (12.7) | 41.7 (17.5) | 60.7 (33.7)a | <0.001 |
| First assessment Trails B | 82.7 (44.4)a | 87.0 (46.9) | 95.9 (45.7)d | 115.7 (63.2)c | 193.2 (86.5)h | <0.001 |
| First assessment LIMM | 14.3 (2.9) | 14.3 (3.3) | 11.0 (2.7) | 7.1 (3.0) | 4.0 (2.6) | <0.001 |
| Race (%) | ||||||
| Asian | 1.26 | 0.00 | 1.51 | 0.72 | 2.91 | |
| Native Hawaiian or Pacific | 0.00 | 0.00 | 0.38 | 0.72 | 0.00 | |
| Black or African American | 8.81 | 1.39 | 1.51 | 2.88 | 2.91 | |
| American Indian or Alaskan | 0.00 | 0.00 | 0.38 | 0.00 | 0.00 | 0.2 |
| White | 88.68 | 95.83 | 93.96 | 94.96 | 93.20 | |
| More than one race | 1.26 | 2.78 | 1.51 | 0.72 | 0.97 | |
| Race unknown | 0.00 | 0.00 | 0.75 | 0.00 | 0.00 | |
| MRI follow-up time from baseline for whole brain atrophy rates (years) | 2.4 (1.3) | 1.7 (0.8) | 2.4 (1.3) | 2.2 (1.3) | 1.0 (0.5) | Not appropriate |
| Number of MRI visits for whole brain atrophy | 4.7 (1.2) | 2.8 (0.9) | 4.4 (1.2) | 4.5 (1.0) | 3.4 (1.0) | Not appropriate |
| Baseline WMH (ml) | 3.5 (4.9) | 3.6 (4.1) | 3.9 (6.9) | 3.6 (6.3) | 6.1 (8.9) | 0.01 |
| Lacunes n (%) | 3 (1.9)b | 1 (1.4)a | 11 (4.2)b | 0 (0.0)a | 2 (2.0)a | 0.1 |
| CMB (1 or more) n (%) | 27 (17.8)f | 6 (8.6)b | 36 (14.0)g | 17 (12.4)b | 21 (20.8)b | 0.2 |
| Whole-brain volume, ml | 1068 (105) | 1094 (88) | 1083 (108) | 1067 (101) | 1027 (117) | <0.001 |
| Total (left plus right) hippocampal volume, ml | 5.46 (0.65) | 5.68 (0.70)a | 5.43 (0.73)e | 5.09 (0.83) | 4.60 (0.71) | <0.001 |
Demographics are shown for controls, Early Mild Cognitive Impairment (EMCI), Late Mild Cognitive Impairment (LMCI), Subjective Memory Concern (SMC) and Alzheimer’s disease. Values are mean (SD) unless stated in the table, White matter hyperintensity (WMH) is reported as median (interquartile range). P-values represent the result of a single test over all groups.
LIMM, logical memory immediate recall score; MMSE, Mini-Mental State Examination.
Missing in 1 subject.
Missing in 2 subjects.
Missing in 3 subjects.
Missing in 4 subjects.
Missing in 5 subjects.
Missing in 7 subjects.
Missing in 8 subjects.
Missing in 12 subjects.
Imaging measures at baseline
Alzheimer’s disease participants had, on average, significantly greater WMH volume than other groups. There was a low prevalence of lacunes (2% overall) and prevalence of CMBs was 15%. There were no significant differences in proportions of subjects with a CMB or lacune across subject groups. Brain and hippocampal volumes differed across groups with volumes in the Alzheimer’s disease group being the smallest.
Atrophy rates
Atrophy rates across the diagnostic groups
All groups experienced significant brain and hippocampal volume loss over time (see Table 2). The fully adjusted model indicated that controls experienced brain atrophy at an average rate of 5.3 ml/year and hippocampal atrophy at 0.04 ml/year, for average TIV and log2WMH, and with no lacunes, and CMBs. Atrophy rates in SMC were not significantly different from controls. Atrophy rates were significantly higher in EMCI patients than controls, with an average whole-brain atrophy rate of 6.8 ml/year, and a hippocampal atrophy rate of 0.06 ml/year. LMCI patients also had a greater mean adjusted atrophy rates than controls, at 9.3 ml/year for the whole brain and 0.11 ml/year for the hippocampus. Individuals with Alzheimer’s disease had the highest mean-adjusted whole-brain atrophy rate (14.7 ml/year) and hippocampal atrophy rates (0.19 ml/year).
Table 2.
Results of the models of brain and hippocampal atrophy rate
| a) Whole-brain atrophy rate, ml/year | b) Hippocampal atrophy rate, ml/year | |||
|---|---|---|---|---|
| Control | 5.3 [4.7, 5.9] | 0.044 [0.036, 0.053] | ||
| SMC | 5.3 [4.3, 6.3] | 0.039 [0.019, 0.059] | ||
| EMCI | 6.8** [6.2, 7.4] | 0.058* [0.048, 0.069] | ||
| LMCI | 9.3** [8.0, 10.5] | 0.113** [0.094, 0.132] | ||
| Alzheimer’s disease | 14.7** [12.9, 16.4] | 0.185** [0.155, 0.215] | ||
|
| ||||
| Change in atrophy rate | Individual models | Adjusted model | Individual models | Adjusted model |
|
| ||||
| for a doubling of WMH volume | 0.3 (0.01) [0.1, 0.6] | 0.3 (0.03) [0.0, 0.6] | 0.013 (<0.001) [0.009, 0.017] | 0.013 (<0.001) [0.008, 0.017] |
| for one or more CMBs | 1.5 (0.004) [0.5, 2.5] | 1.4 (0.007) [0.4, 2.4] | 0.029 (0.001) [0.013, 0.045] | 0.021 (0.009) [0.005, 0.04] |
The top half of the table represents mean [95% CI] atrophy rates in mls per year are shown for the 5 diagnostic groups. The atrophy rates are the mean predicted rate of change per year in each group calculated from the mixed model. The bottom half of the table shows the individual effects of WMH and CMBs on atrophy rates together with mutually adjusted associations that are also adjusted for lacunes. Results here are changes in atrophy rate for given increases in SVD, (P-value), [95% CI]. All models are adjusted for total intracranial volume. Values in the bottom half of the table are adjusted for diagnostic group.
EMCI, Early Mild Cognitive Impairment; LMCI, Late Mild Cognitive Impairment; SMC, Subjective Memory Concern; SVD, small vessel disease; WMH, white matter hyperintensity.
P-value represents a difference in rates from controls at P < 0.05.
Significantly different from controls P < 0.01.
SVD predicting atrophy rates
WMHs and CMBs separately and independently predicted hippocampal and whole-brain atrophy rate across all individuals adjusted for diagnostic group (see Table 2). For both of these predictors, an increase in WMH or presence of CMB was associated with greater volume loss. A doubling of baseline WMH volume was associated with an increase in whole-brain atrophy rates of 0.3 ml/year (95% CI 0.1, 0.6) and an increase in hippocampal atrophy of 0.013 ml/year (95% CI 0.009, 0.017). Presence of a least one CMB, compared with those without CMB, was associated with an increase in 1.5 ml/year (95% CI 0.5, 2.5) whole-brain atrophy and an increase in hippocampal atrophy of 0.029 ml/year (95% CI 0.013, 0.045). There was no evidence that the association of WMH with atrophy rate differed according to diagnostic group (P = 0.62 for whole-brain and P = 0.70 for hippocampal atrophy rate) or that the association of CMBs differed by diagnostic group (P = 0.12 for whole brain and P = 0.18 for hippocampal atrophy). There was no evidence to suggest the association between WMH and atrophy rates differed in those with and without a CMB for either the whole brain (P = 0.14) or the hippocampus (P = 0.38) over all participants.
Neuropsychology
Neuropsychological changes across the diagnostic groups
Table 3 shows the change in neuropsychology results and the associations between SVD measures and these changes. Controls saw either no evidence of change in neuropsychology measures over time (MMSE, Trails A and B) or a modest improvement over time [LIMM, 0.23; (95% CI 0.09, 0.37) points/year]. SMC saw no significant changes over time. EMCI only showed significant decline in MMSE over time −0.21 (95% CI −0.32, −0.12). LMCI and Alzheimer’s disease tended to show decline on all measures, but decline was statistically significant only for the LMCI group for MMSE and Trails B and for Alzheimer’s disease for MMSE and Trails A. For MMSE, this was a decline of −1.01 points per year for LMCI (95% CI −1.30, −0.78) and −2.34 points per year for Alzheimer’s disease (95% CI −3.13, −1.74). Significant differences were seen between controls and EMCI for MMSE, LMCI for LIMM and MMSE and between controls and Alzheimer’s disease in MMSE, LIMM, and Trails A.
Table 3.
Results of the models of change in neuropsychology measures
| MMSE | LIMM | Trails A | Trails B | |||||
|---|---|---|---|---|---|---|---|---|
| Control | −0.06 [−0.16, 0.02] | 0.23 [0.09, 0.37] | 0.01 [−0.03, 0.05] | −0.01 [−0.04, 0.01] | ||||
| SMC | −0.10 [−0.25, 0.01] | 0.14 [−0.21, 0.48] | 0.04 [−0.04, 0.13] | −0.00 [−0.04, 0.04] | ||||
| EMCI | −0.21 [−0.32, −0.12]* | 0.18 [0.05, 0.32] | −0.01 [−0.04, 0.02] | −0.00 [−0.02, 0.01] | ||||
| LMCI | −1.01 [−1.30, −0.78]* | −0.09 [−0.29, 0.10]** | −0.04 [−0.09. 0.00] | −0.04 [−0.06, −0.02] | ||||
| Alzheimer’s disease | −2.34 [−3.13, −1.74]* | −0.25 [−0.66, 0.16]* | −0.22 [−0.35, −0.09]** | −0.05 [−0.11, 0.02] | ||||
|
| ||||||||
| Difference in change in cognition | Individual models | Adjusted models | Individual models | Adjusted models | Individual models | Adjusted models | Individual models | Adjusted models |
|
| ||||||||
| for a doubling of WMH volume | −0.06 [−0.11, −0.02] | −0.06 [−0.10, −0.00] | −0.11 (<0.001) [−0.16, −0.05] | −0.09 (0.001) [−0.15, −0.04] | −0.02 (0.009) [−0.03, −0.004] | −0.02 (0.005) [−0.03, −0.01] | −0.01 (0.05) [−0.01, −0.00] | −0.01 (0.04) [−0.01, −0.00] |
| for one or more CMBs | −0.25 [−0.56, −0.06] | −0.21 [−0.50, −0.02] | −0.42 (<0.001) [−0.66, −0.19] | −0.32 (0.006) [−0.55, −0.09] | −0.04 (0.15) [−0.10, 0.01] | −0.03 (0.34) [−0.08, 0.03] | −0.01 (0.35) [−0.04, 0.01] | −0.01 (0.60) [−0.04, 0.02] |
Mean [95% CI] change in score per year are shown for the 5 diagnostic groups in the top of the table. Of note, the Trails A and Trails B scores are transformed (see Methods section). The bottom half of the table shows the individual effects of WMH and CMB on change in neuropsychology scores together with the mutually adjusted associations that are also adjusted for lacunes. Results here are changes in neuropsychology test score for given increases in SVD, (P-value), [95% CI]. Of note, no P-value is given for the MMSE result; significance is determined by 95% CI not including 0.
EMCI, Early Mild Cognitive Impairment; LIMM, logical memory immediate recall score; LMCI, Late Mild Cognitive Impairment; MMSE, Mini-Mental State Examination; SMC, Subjective Memory Concern; SVD, small vessel disease; WMH, white matter hyperintensity.
P-value represents a difference in rates from controls at P < 0.05.
Significantly different from controls P < 0.01.
SVD predicting neuropsychology
Across all individuals and adjusted for diagnostic group, greater WMH volume was associated with greater cognitive decline. A doubling of baseline WMH was associated with a reduction in change for: MMSE of −0.06 points/year (95% CI, −0.10, −0.00); LIMM of −0.09 points/year (95% CI −0.15, −0.04); Trails A of −0.02 100/seconds per year (95% CI −0.03, −0.01); Trails B of −0.01 100/seconds per year (95% CI −0.01, −0.00) (Table 3). Presence of one or more CMB, compared with no CMB, was associated with greater decline in MMSE by −0.21 points/year (95% CI −0.50, −0.02) and LIMM by −0.32 points/year (95% CI −0.55, −0.09) adjusting for WMH and lacunes, but there was little evidence of associations with changes in Trails A or Trails B. There was no evidence WMH or CMB had a differential effect by diagnostic group on change over time in any of the neuropsychology outcomes (P > 0.3, all tests). There was no evidence of an interaction between CMB and WMH for any neuropsychological test outcome (P > 0.2, all tests).
Disease-progression measure modelling
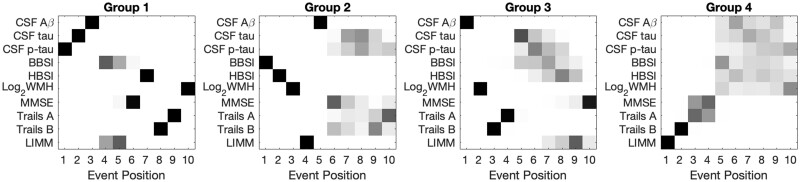
Four hundred and fifty individuals had complete datasets that were used in the SuStaIn EBM and cross-cluster group analyses. Figure 3 and Table 4 show four different groups of individuals identified from this technique, and Table 5 shows summary statistics for the biomarkers used in SuStaIn EBM by group.
Figure 3.
Results from disease progression modelling analyses. Subject numbers for each group are: Group 1 (223); Group 2 (108); Group 3 (76); Group 4 (43). Only continuous complete-case biomarker variables were used. No time scale is imposed on the event position (x-axis); events may be close together or distant in time. Positioning for all biomarkers of interest (y-axis) in all groups is presented. Each entry of each positional variance diagram corresponds to the probability each biomarker (y-axis) will become abnormal at each position in the sequence (x-axis) estimated by the SuStaIn EBM algorithm, ranging from 0 in white to 1 in black. Definitions: cerebrospinal fluid (CSF); amyloid beta 1–42 (Aβ); total tau (tau); phosphorylated tau 181 (p-tau); brain boundary shift integral atrophy rate over 12 months (BBSI); hippocampal boundary shift integral atrophy rate over 12 months (HBSI); Mini Mental State Examination (MMSE); logical memory immediate recall (LIMM).
Table 4.
Analysis of cross-sectional and longitudinal variables of interest that were not entered into the SuStaIn EBM algorithm according to groups (clusters) derived from the SuStaIn EBM algorithm
| Group 1 223 | Group 2 108 | Group 3 76 | Group 4 43 | P-value across groups | |
|---|---|---|---|---|---|
| Diagnosis % C/SMC/EMCI/LMCI/Alzheimer’s disease | 26/3/37/24/10 | 21/6/44/17/12 | 37/3/46/14/0 | 2/0/44/42/12 | <0.001 |
| Stage % 0–1/2–3/4–5/6–7/8–10 | 47/14/10/13/17 | 36/20/21/10/12 | 33/32/14/3/18 | 42/26/26/2/5 | <0.001 |
| Age at baseline, years | 71.2 (7.0) | 72.5 (7.2) | 75.8 (6.2) | 71.4 (8.5) | <0.001 |
| Male % | 48 | 65 | 54 | 44 | 0.018 |
| Education, years | 16.2 (2.5) | 16.6 (2.8) | 16.4 (2.4) | 15.6 (2.7) | 0.31a |
| History of hypertension % | 44 | 44 | 71 | 44 | <0.001 |
| Smoking never/previously/current % | 60/39/1 | 61/32/6 | 55/43/1 | 63/35/2 | 0.13 |
| Body Mass Index, kg/m2 | 27.3 (4.6) | 27.7 (5.0)c | 26.8 (5.0) | 29.3 (6.5) | 0.046 |
| Percentage with diabetes | 9 | 10 | 7 | 16 | 0.38 |
| APOE ε4 carrier % | 49 | 34 | 46 | 28 | 0.010 |
| Geriatric depression score (GDS) | 1.4 (1.4) | 1.3 (1.3) | 1.2 (1.5) | 1.9 (1.7) | 0.06 |
| MMSE (pt/year) | −0.4 (2.0) | −0.3 (2.1) | −0.7 (1.5) | −0.2 (1.7) | 0.41 |
| Trails A (pt/year) | 2.5 (14.4) | 2.6 (13.8) | −0.1 (12.9) | −0.4 (13.0) | 0.33 |
| Trails B (pt/year) | 10.9 (37.6) | 10.9 (49.5) | 6.9 (51.9) | −1.6 (39.2) | 0.34 |
| LIMM (pt/year) | 0.4 (3.0) | 1.0 (3.0) | 0.9 (2.8) | 1.7 (3.2) | 0.03 |
| Microbleeds % at least 1 | 11 | 19 | 26 | 7 | 0.004 |
| Brain volume, ml | 1064 (107) | 1087 (107) | 1072 (96) | 1065 (106) | 0.44b |
| Hippocampal volume, ml | 5.22 (0.82) | 5.36 (0.72) | 5.32 (0.66) | 5.23 (0.88) | 0.27b |
EMCI, Early Mild Cognitive Impairment; LIMM, logical memory immediate recall score; LMCI, Late Mild Cognitive Impairment; MMSE, Mini-Mental State Examination; SMC, Subjective Memory Concern; WMH, white matter hyperintensity.
Adjusted for gender.
Adjusted for TIV, age and gender.
Data missing in one individual.
Table 5.
Mean (SD) values of variables used in SuStaIn EBM according to groups (clusters) derived from the SuStaIn EBM algorithm
| Group 1 223 | Group 2 108 | Group 3 76 | Group 4 43 | |
|---|---|---|---|---|
| Aβ_1–42, pg/ml | 247.0 (83.9) | 271.5 (88.8) | 182.7 (44.1) | 300.7 (65.6) |
| Total Tau, pg/ml | 92.5 (49.9) | 65.0 (33.4) | 68.7 (32.7) | 54.0 (18.9) |
| Ptau_181, pg/ml | 28.7 (13.8) | 20.6 (10.6) | 25.0 (10.7) | 19.1 (6.2) |
| BBSI, mls/year | 6.7 (8.2) | 12.5 (5.2) | 6.6 (7.1) | 4.0 (7.1) |
| HBSI, mls/year | 0.07 (0.11) | 0.13 (0.12) | 0.06 (0.10) | 0.02 (0.10) |
| log2WMH, ml | 11.3 (1.2) | 12.5 (1.5) | 12.7 (1.4) | 11.5 (1.2) |
| MMSE,/30 | 27.8 (2.2) | 27.7 (2.3) | 29.0 (1.0) | 27.3 (2.6) |
| TrailsA,/150 | 36.1 (16.3) | 33.8 (10.2) | 41.8 (14.8) | 43.5 (19.7) |
| TrailsB,/300 | 94.5 (55.7) | 90.6 (39.7) | 117.4 (64.2) | 120.5 (61.4) |
| LIMM,/25 | 10.7 (4.4) | 10.1 (4.3) | 12.2 (3.5) | 7.5 (2.7) |
All values are derived from baseline measures apart from the BBSI and HBSI which was calculated from the baseline and 12-month interval scans.
BBSI, Brain Boundary Shift Integral; HBSI, Hippocampal Boundary Shift Integral; LIMM, logical memory immediate recall score; MMSE, Mini-Mental State Examination; WMH, white matter hyperintensity.
Group 1 shows a sequence with ptau, tau and the Aβ 1–42 as initial events, followed by whole-brain atrophy rates (BBSI) and LIMM, then MMSE and hippocampal atrophy rates (HBSI), the Trails tests and finally WMH.
Group 2 has whole-brain atrophy rate (BBSI) as the first event, followed by hippocampal atrophy rate (HBSI) and WMH, then LIMM, Aβ 1–42, and then MMSE, tau, ptau and the other psychological measures.
Group 3 has Aβ 1–42 as the first event, then WMH, followed by Trails B then A, then both tau and ptau and atrophy rate measures (BBSI and HBSI), and finally LIMM and MMSE.
Group 4 shows LIMM as the first event, then Trails B, Trails A and MMSE and then all other measures in a block of uncertain ordering.
In terms of other characteristics, significant differences across the groups were shown according to proportions of diagnostic groups, SuStaIn stage, baseline age, gender, hypertension history, APOEε4 status, change in LIMM, and presence of at least one CMB (see Table 4). Notable features of Group 1, the largest group of 223 individuals, were that this group had the lowest proportion of EMCI, the highest proportion of the lowest SuStaIn stage category and APOE ε4 carriers (49%) and the lowest LIMM improvement scores (0.4 points/year). Group 2 had the highest proportion of men (65%) and a distribution of proportions of diagnoses similar to Group 1 albeit with the highest proportion of SMCs (6%) and the joint highest proportion of Alzheimer’s disease (12%). For Group 3, this had the highest proportion of controls (37%) and EMCI (46%), the lowest proportion of LMCI (14%) and no Alzheimer’s disease subjects. This group had the highest proportion of the most severe SuStaIn stage category (18%). This was the oldest group at 75.8 years with the highest history of hypertension (71%) and microbleeds (26%) and the lowest BMI. Group 4 was the smallest group (43 individuals) with the highest proportion of LMCI (42%) and the joint-highest proportion of Alzheimer’s disease (12%), the lowest proportion of controls (2%) and no SMC and the lowest proportion of the most severe SuStaIn stage category (5%). This group had the highest change in LIMM (1.7 points per year improvement), the lowest proportion of men (44%), and the highest BMI.
We investigated baseline depression scores over the four groups and found borderline evidence in differences across groups with slightly higher mean scores in Group 4 (P = 0.06). We also investigated conversion status during the 12-month period from baseline. Conversion from MCI to dementia occurred at rates of 5, 7, 3 and 5% for Groups, 1, 2, 3 and 4, respectively. From Group 1, 1% converted from normal cognition to MCI and 3% from Groups 2 and 3. Reversion from MCI to normal cognition after 12 months occurred at rates of 2, 1, 5 and 7% for Groups 1, 2, 3 and 4, respectively.
Discussion
This study provides evidence that presumed SVD markers are associated with measures of disease progression. We found that baseline WMH volume and CMB presence were independently associated with increased whole-brain and hippocampal atrophy rate and decline in general cognition and logical memory performance. We established four distinct groups with different biomarker sequences with WMH being an early event for two groups and a later event for two others.
The four groups identified by disease-progression modelling potentially represent different phenotypes in ADNI2/Go. Group 1 individuals, about half of those analysed, may be on a typical Alzheimer’s disease pathway with CSF tau and amyloid measures being the first biomarkers becoming abnormal. These individuals had an average CSF amyloid of 247 pg/ml which indicates amyloidosis (the cut-point has been reported as 254 pg/ml) and the highest tau and ptau levels. In this group, WMH development was a later event following amyloid and tau pathology and neurodegeneration. WMH can be caused by pathologies other than cerebrovascular disease and WMH here may be the result of neurodegeneration or classical Alzheimer’s disease pathologies.29,30 Group 2, with early neurodegenerative markers, could be a ‘suspected non-Alzheimer’s disease pathophysiology’31 or ‘non-Alzheimer’s disease pathological change’ group.5 This group had the highest atrophy rates and relatively high WMH values but without cerebral amyloidosis on average. The third and oldest group, which was the least overweight, showed more mixed pathology with amyloidosis and WMH being early events. The average CSF amyloid level was the lowest of all groups. This group may represent those with some cerebral amyloid angiopathy (CAA) since they had the highest CMB proportion. The fourth and smallest group, showed early cross-sectional neuropsychological score abnormalities with little certainty regarding imaging and CSF measure ordering. This group improved most on logical memory test over one year but had the lowest baseline scores. These individuals had the lowest proportion in the most severe SuStaIn stage category and were most overweight. This group may represent those who may, on average, have temporary impairments.
Our findings, from the disease-progression modelling results in Group 3 in particular, are similar to another publication which shows vascular dysfunction as an early disease event.32 Our work differs as we assessed WMH and CMB as well as progression heterogeneity. Similar work in a different cohort established four clusters which approximated to: preclinical typical Alzheimer’s disease; mixed vascular and preclinical Alzheimer’s disease; atrophy-based; typical ageing.33 Other research showed different patterns of biomarker evolution in amyloid positive versus negative individuals from ADNI1.34 This approach is similar to ours as different patterns of disease progression are expected. Our work differs by including SVD markers and allowing groups to be determined by the data Another study assessed clusters using baseline MRI, CSF and serum biomarkers in ADNI1 MCI subjects.35 That study found considerable heterogeneity in the MCI population with four different clusters that approximated to: controls; Alzheimer’s diseases; and two early Alzheimer’s disease groups. One of these early Alzheimer’s disease groups did not seem to conform to the expected progression models suggesting that not all individuals are on the same trajectory. Our work extends this by including all newly enrolled ADNI2/Go subjects and including presumed cerebrovascular, CSF, atrophy and cognitive scores.
The atrophy rate results are in keeping with the literature.36–38 Approximate whole-brain atrophy rates as % baseline volume per year were controls: 0.5; SMC: 0.5; EMCI: 0.6; LMCI 0.9; Alzheimer’s disease: 1.4. Analogous hippocampal rates were controls: 0.7; SMC 0.7; EMCI: 1.1; LMCI 2.2; Alzheimer’s disease: 4.1.
This work confirms that greater WMH volumes are associated with subsequent brain volume decline, as shown previously.6–8,39 Converting our reported effects from ml/year to %/year of baseline volume, a doubling of WMH was associated with an approximate increase of 0.03%/year for brain atrophy rates, and 0.25%/year for hippocampal atrophy rates. Others have found that greater WMH volumes are associated with reductions in temporal lobe volumes in MCI.40–43 Our work extends this by reporting that the relationship between WMHs and progressive atrophy is independent of other markers of presumed SVD. A novel finding of this study is that CMB presence is associated with higher brain and hippocampal atrophy rates. The effect size was large: presence of a CMB associated with an increase in atrophy rate equivalent to approximately 25% of the atrophy rate in controls without a CMB. Two studies have found associations between CMBs and cross-sectional measures of brain atrophy.44,45 One study found no evidence for a relationship between newly developing CMBs and brain volume change.9 Atrophy rate being related to CMBs may be driven by underlying advanced Alzheimer’s disease pathology causing CAA and micro-haemorrhage. The CMB prevalence in this study was 15%, which is similar to other comparably aged groups: 11% and 31%46,47; although our study had a higher field-strength scanner compared with these studies (3 T compared with 1.5 T) the latter used a thinner-slice gradient-echo technique. Furthermore, our study did not include those with a high vascular disease burden.
Our finding that WMHs are associated with neuropsychological decline is in keeping with previous findings48,49 including from this cohort.50 This work extends the literature by showing the WMH-cognitive decline relationship to be independent of CMBs and lacunes. Presence of a CMB was also associated with decline in logical memory which was also independent of lacunes and WMH. This is broadly in line with results showing newly developing CMBs are associated with decline.51 However, although a recent review suggested that CMBs affected global cognitive performance, it concluded that CMBs mainly affect executive function which we did not find.49
A strength of this work is that we used multiple methodologies to investigate relationships between SVD and Alzheimer’s disease biomarkers. However, limitations include that this dataset was select; ADNI participants are in relatively good health without severe vascular disease. We did not look at the effect of number or locations of CMB, but assumed global effects on outcomes for this first analysis. Location of SVD is an important topic for future research since those with lobar CMB may have different associations with progression measures compared with those with deep/basal ganglia CMBs. Mis-segmentation of WMH does occur with automated algorithms and semi-automatically, or manually, segmenting WMH may reveal different results, however, the algorithm used has shown good comparability compared with semi-automated (guided-manual) segmentations50 and segmentations were visually inspected. Importantly, follow-up in the Alzheimer’s disease group was shorter which limits the extent to which changes can be detected in this group; caution is needed when interpreting between-group differences. Furthermore, participants with vascular lesions in the early stages of disease (controls, SMC, EMCI), may not be on the same mechanistic pathway to cognitive decline and dementia as LMCI and Alzheimer’s disease groups. A consideration is that we did not adjust for age when assessing the effects of SVD on neuropsychological scores or atrophy rates. We chose this approach since age and SVD are strongly associated and age may act as a proxy for the accumulation of vascular disease and interact with processes along the causal pathway. We also did not investigate the effects of: education; APOE ε4; disease severity; disease length on outcomes. We found a very low prevalence of lacunes (2%) and therefore did not present the lacune effects on outcomes. This low prevalence may be in part due to the fact that we only assessed lacunes in the white matter. We did not have complete data on all subjects to include in the SuStaIn EBM and this may have influenced the findings of this approach. We did not transform the neuropsychological scores used in the SuStaIn EBM models since there was no adequate transform for MMSE. Further work using groupings derived from data-driven approaches will be important. For example, investigating progression and eventual pathological examination of brain tissue according to grouping may reveal differences in outcomes and underlying (co)pathologies. Notably, recent work has suggested that WMHs are not always associated with vascular disease at post mortem30. Autopsy confirmation of diagnosis was not available in this study.
In summary, WMHs and CMBs are independently associated with brain and hippocampal volume change. This suggests that CMBs and WMHs have independent mechanisms which may contribute to neurodegeneration. WMH may be an early event for groups that either show suspected non-Alzheimer’s disease pathology, or older subjects that may have mixed Alzheimer’s and cerebrovascular disease or CAA. Markers of presumed SVD are important to consider when assessing decline in non-demented and clinical Alzheimer’s disease subjects. The heterogeneity found in this cohort suggests that different clinical approaches may be needed for different groups of individuals.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
The authors would like to thank Duncan Wilson and Rachel Sparks for their expertise when performing image analysis.
Funding
This work was supported by an Alzheimer’s Research United Kingdom Senior Research Fellowship (J.B. and C.F.). C.H.S. is supported by an Alzheimer’s Society Junior Fellowship (AS-JF-17-011). P.W. is supported by a Wolfson Foundation studentship. This work is supported by the National Institute Health Research University College London/Hospital Biomedical Research Centre which also specifically supports O.G. and F.B. D.A. and A.L.Y. received support from Engineering and Physical Sciences Research Council grants EP/J020990/01 and EP/M020533/1 and the European Union’s Horizon 2020 research and innovation programme under grant agreement No 666992 (EuroPOND). A.L.Y. is also supported by a Medical Research Council Skills Development Fellowship. This work has also been supported by a Wellcome/Engineering and Physical Sciences Research Council Centre for Medical Engineering (WT203148/Z/16/Z) and Wellcome Flagship Programme (WT213038/Z/18/Z). I.M. is supported by the following grants: Alzheimer’s Research United Kingdom (ARUK-PG2014-1946, ARUK-PG2017-1946), and the Wolfson Foundation (PR/ylr/18575).
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org Accessed 07 October 2021). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Competing interests
Prof Barkhof reports being on the board of Neurology, Brain, Radiology, MSJ, and being section editor of Neuroradiology. He has received personal fees from Springer, Bayer, IXICO Ltd, Biogen, Roche, GeNeuro. He has had grants from Novartis, TEVA, Merck, Biogen, Innovative Medicines Initiative—European Union, General Electric Healthcare, United Kingdom Multiple Sclerosis Society, Dutch Foundation Multiple Sclerosis Research, The Dutch Research Council (NOW), National Institute Health Research. These are outside the submitted work. Prof Alexander is an associate editor of Magnetic Resonance in Imaging and of Medical Image Analysis and on the editorial board for Neuroimage. Dr Barnes is an associate editor of Journal of Alzheimer’s Disease. Hugh Pemberton took a position at GE healthcare following submission of this manuscript.
Glossary
- Aβ =
amyloid beta
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- APOE =
apolipoprotein E
- BaMoS =
Bayesian model selection
- BBSI =
brain boundary shift integral
- BMI =
body mass index
- BSI =
boundary shift integral
- CAA =
cerebral amyloid angiopathy
- CDR =
clinical dementia rating
- CMBs =
cerebral microbleeds
- EBM =
event-based model
- EMCI =
early mild cognitive impairment
- FLAIR =
fluid-attenuated inversion recovery
- GIF =
geodesic information flows
- HBSI =
hippocampal boundary shift integral
- LIMM =
logical memory—immediate recall
- LMCI =
late mild cognitive impairment
- MAPS =
multi-atlas propagation and segmentation
- MCI =
mild cognitive impairment
- MCMC =
Markov Chain Monte Carlo
- MMSE =
Mini-Mental State Examination
- NINCDS/ADRDA =
National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association
- ptau =
phosphorylated tau
- SMC =
significant memory concern
- SPM12 =
statistical parametric mapping
- STEPS =
similarity and truth estimation for propagated segmentations
- SuStaIn =
Subtype and Stage Inference
- SVD =
small vessel disease
- TIV =
total intracranial volume
- WMHs =
white matter hyperintensities
References
- 1. Vermeer SE, Longstreth WT, Koudstaal PJ.. Silent brain infarcts: A systematic review. Lancet Neurol. 2007;6(7):611–619. [DOI] [PubMed] [Google Scholar]
- 2. Jeerakathil T, Wolf PA, Beiser A, et al. Cerebral microbleeds: Prevalence and associations with cardiovascular risk factors in the Framingham Study. Stroke. 2004;35(8):1831–1835. [DOI] [PubMed] [Google Scholar]
- 3. Yoshita M, Fletcher E, Harvey D, et al. Extent and distribution of white matter hyperintensities in normal aging, MCI, and AD. Neurology. 2006;67(12):2192–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wardlaw JM, Smith EE, Biessels GJ, et al. ; STandards for ReportIng Vascular changes on nEuroimaging (STRIVE v1). Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12(8):822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jack CR, Bennett DA, Blennow K, et al. ; Contributors. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barnes J, Carmichael OT, Leung KK, et al. Vascular and Alzheimer’s disease markers independently predict brain atrophy rate in Alzheimer’s Disease Neuroimaging Initiative controls. Neurobiol Aging. 2013;34(8):1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Enzinger C, Fazekas F, Matthews PM, et al. Risk factors for progression of brain atrophy in aging: Six-year follow-up of normal subjects. Neurology. 2005;64(10):1704–1711. [DOI] [PubMed] [Google Scholar]
- 8. Fiford CM, Manning EN, Bartlett JW, et al. ; Alzheimer's Disease Neuroimaging Initiative. White matter hyperintensities are associated with disproportionate progressive hippocampal atrophy. Hippocampus. 2017;27(3):249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goos JDC, Henneman WJP, Sluimer JD, et al. Incidence of cerebral microbleeds: A longitudinal study in a memory clinic population. Neurology. 2010;74(24):1954–1960. [DOI] [PubMed] [Google Scholar]
- 10. Young AL, Oxtoby NP, Daga P, et al. A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain. 2014;137(9):2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hachinski VC, Iliff LD, Zilhka E, et al. Cerebral blood flow in dementia. Arch Neurol. 1975;32(9):632–637. [DOI] [PubMed] [Google Scholar]
- 12. Manning EN, Leung KK, Nicholas JM, et al. ; Alzheimer’s Disease Neuroimaging Initiative. A comparison of accelerated and non-accelerated MRI scans for brain volume and boundary shift integral measures of volume change: Evidence from the ADNI dataset. Neuroinformatics. 2017;15(2):215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jovicich J, Czanner S, Greve D, et al. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30(2):436–443. [DOI] [PubMed] [Google Scholar]
- 14. Sled JG, Zijdenbos AP, Evans AC.. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17(1):87–97. [DOI] [PubMed] [Google Scholar]
- 15. Sudre CH, Cardoso MJ, Bouvy WH, Biessels GJ, Barnes J, Ourselin S.. Bayesian model selection for pathological neuroimaging data applied to white matter lesion segmentation. IEEE Trans Med Imaging. 2015;34(10):2079–2102. [DOI] [PubMed] [Google Scholar]
- 16. Cardoso MJ, Modat M, Wolz R, et al. Geodesic information flows: Spatially-variant graphs and their application to segmentation and fusion. IEEE Trans Med Imaging. 2015;34(9):1976–1988. [DOI] [PubMed] [Google Scholar]
- 17. Modat M, Cash DM, Daga P, Winston GP, Duncan JS, Ourselin S.. Global image registration using a symmetric block-matching approach. J Med Imaging. 2014;1(2):024003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clarkson MJ, Zombori G, Thompson S, et al. The NifTK software platform for image-guided interventions: Platform overview and NiftyLink messaging. Int J Comput Assist Radiol Surg. 2015;10(3):301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gregoire SM, Chaudhary UJ, Brown MM, et al. The Microbleed Anatomical Rating Scale (MARS): Reliability of a tool to map brain microbleeds. Neurology. 2009;73(21):1759–1766. [DOI] [PubMed] [Google Scholar]
- 20. Leung KK, Barnes J, Modat M, et al. Brain MAPS: An automated, accurate and robust brain extraction technique using a template library. Neuroimage. 2011;55(3):1091–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cardoso MJ, Leung K, Modat M, et al. STEPS: Similarity and Truth Estimation for Propagated Segmentations and its application to hippocampal segmentation and brain parcelation. Med Image Anal. 2013;17(6):671–684. [DOI] [PubMed] [Google Scholar]
- 22. Leung KK, Ridgway GR, Ourselin S, Fox NC;. Alzheimer's Disease Neuroimaging Initiative. Consistent multi-time-point brain atrophy estimation from the boundary shift integral. Neuroimage. 2012;59(4):3995–4005. [DOI] [PubMed] [Google Scholar]
- 23. Malone IB, Leung KK, Clegg S, et al. Accurate automatic estimation of total intracranial volume: A nuisance variable with less nuisance. Neuroimage. 2015;104:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barnes J, Ridgway GR, Bartlett J, et al. Head size, age and gender adjustment in MRI studies: A necessary nuisance? Neuroimage. 2010;53(4):1244–1255. [DOI] [PubMed] [Google Scholar]
- 25. Weintraub S, Salmon D, Mercaldo N, et al. The Alzheimer’s Disease Centers’ Uniform Data Set (UDS): The Neuropsychologic Test Battery. Alzheimer Dis Assoc Disord. 2009;23(2):91–101. https://www.alz.washington.edu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frost C, Kenward MG, Fox NC.. The analysis of repeated “direct” measures of change illustrated with an application in longitudinal imaging. Stat Med. 2004;23(21):3275–3286. [DOI] [PubMed] [Google Scholar]
- 27. Young AL, Marinescu RV, Oxtoby NP, et al. ; The Genetic FTD Initiative (GENFI). Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat Commun. 2018;9(1):1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fonteijn HM, Modat M, Clarkson MJ, et al. An event-based model for disease progression and its application in familial Alzheimer’s disease and Huntington’s disease. Neuroimage. 2012;60(3):1880–1889. [DOI] [PubMed] [Google Scholar]
- 29. McAleese KE, Firbank M, Dey M, et al. Cortical tau load is associated with white matter hyperintensities. Acta Neuropathol Commun. 2015;3:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McAleese KE, Walker L, Graham S, et al. Parietal white matter lesions in Alzheimer’s disease are associated with cortical neurodegenerative pathology, but not with small vessel disease. Acta Neuropathol. 2017;134(3):459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jack CR, Knopman DS, Weigand SD, et al. An operational approach to National Institute on Aging-Alzheimer’s Association criteria for preclinical Alzheimer disease. Ann Neurol. 2012;71(6):765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iturria-Medina Y, Sotero RC, Toussaint PJ, et al. ; Alzheimer’s Disease Neuroimaging Initiative. Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat Commun. 2016;7(May):11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Racine AM, Koscik RL, Berman SE, et al. Biomarker clusters are differentially associated with longitudinal cognitive decline in late midlife. Brain. 2016;139(Pt 8):2261–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bertens D, Knol DL, Scheltens P, Visser PJ; Alzheimer's Disease Neuroimaging Initiative. Temporal evolution of biomarkers and cognitive markers in the asymptomatic, MCI, and dementia stage of Alzheimer’s disease. Alzheimers Dement. 2015;11(5):511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nettiksimmons J, DeCarli C, Landau S, Beckett L; Alzheimer's Disease Neuroimaging Initiative. Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimers Dement. 2014;10(5):511–521.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hua X, Ching CRK, Mezher A, et al. ; Alzheimer's Disease Neuroimaging Initiative. MRI-based brain atrophy rates in ADNI phase 2: Acceleration and enrichment considerations for clinical trials. Neurobiol Aging. 2016;37:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iglesias JE, Van Leemput K, Augustinack J, Insausti R, Fischl B, Reuter M; Alzheimer's Disease Neuroimaging Initiative. Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject-specific atlases. Neuroimage. 2016;141:542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fletcher E, Filshtein TJ, Harvey D, Renaud A, Mungas D, DeCarli C.. Staging of amyloid β, t-tau, regional atrophy rates, and cognitive change in a nondemented cohort: Results of serial mediation analyses. Alzheimers Dement Diagn Assess Dis Monit. 2018;10:382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Guio F, Duering M, Fazekas F, et al. Brain atrophy in cerebral small vessel diseases: Extent, consequences, technical limitations and perspectives: The HARNESS initiative. J Cereb Blood Flow Metab. 2020;40(2):231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eckerström C, Olsson E, Klasson N, et al. High white matter lesion load is associated with hippocampal atrophy in mild cognitive impairment. Dement Geriatr Cogn Disord. 2011;31(2):132–138. [DOI] [PubMed] [Google Scholar]
- 41. Ye BS, Seo SW, Kim GH, et al. Amyloid burden, cerebrovascular disease, brain atrophy, and cognition in cognitively impaired patients. Alzheimers Dement. 2014;11(5):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guzman V, Carmichael OT, Schwarz C, Tosto G, Zimmerman ME, Brickman AM.. White matter hyperintensities and amyloid are independently associated with entorhinal cortex volume among individuals with mild cognitive impairment. Alzheimers Dement. 2013;9(5 Suppl.):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tosto G, Zimmerman ME, Hamilton JL, Carmichael OT, Brickman AM., Alzheimer's Disease Neuroimaging Initiative. The effect of white matter hyperintensities on neurodegeneration in mild cognitive impairment. Alzheimers Dement. 2015;11(12):1510–1519. doi: 10.1016/j.jalz.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chowdhury MH, Nagai A, Bokura H, Nakamura E, Kobayashi S, Yamaguchi S.. Age-related changes in white matter lesions, hippocampal atrophy, and cerebral microbleeds in healthy subjects without major cerebrovascular risk factors. J Stroke Cerebrovasc Dis. 2011;20(4):302–309. [DOI] [PubMed] [Google Scholar]
- 45. Samuraki M, Matsunari I, Yoshita M, et al. Cerebral amyloid angiopathy-related microbleeds correlate with glucose metabolism and brain volume in Alzheimer’s disease. J Alzheimer’s Dis. 2015;48(2):517–528. [DOI] [PubMed] [Google Scholar]
- 46. Sveinbjornsdottir S, Sigurdsson S, Aspelund T, et al. Cerebral microbleeds in the population based AGES-Reykjavik study: Prevalence and location. J Neurol Neurosurg Psychiatry. 2008;79(9):1002–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vernooij M, van der Lugt A, Ikram M, et al. Prevalence and risk factors of cerebral microbleeds. Neurology. 2008;70(14):1208–1214. [DOI] [PubMed] [Google Scholar]
- 48. Carmichael O, Schwarz C, Drucker D, et al. ; Alzheimer's Disease Neuroimaging Initiative. Longitudinal changes in white matter disease and cognition in the first year of the Alzheimer disease neuroimaging initiative. Arch Neurol. 2010;67(11):1370–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caunca MR, De Leon-Benedetti A, Latour L, Leigh R, Wright CB.. Neuroimaging of cerebral small vessel disease and age-related cognitive changes. Front Aging Neurosci. 2019;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fiford CM, Sudre CH, Pemberton H, et al. for the Alzheimer’s Disease Neuroimaging Initiative. Automated white matter hyperintensity segmentation using Bayesian model selection: Assessment and correlations with cognitive change. Neuroinformatics. 2020;18 (3):429- 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Basselerie H, Bracoud L, Zeestraten E, et al. Incident cerebral microbleeds detected by susceptibility weight-imaging help to identify patients with mild cognitive impairment progressing to Alzheimer’s disease. J Alzheimer’s Dis. 2017;60(1):253–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADNI data are available for download for approved researchers (www.adni-info.org Accessed 07 October 2021). The NiftyMidas visualization and labelling software is available (https://github.com/NifTK/NifTK Accessed 07 October 2021). NiftyReg (used for the linear registration) is available (https://github.com/KCL-BMEIS/niftyreg/wiki Accessed 07 October 2021).