Figure 3.
Prolonged culture expansion selects against putatively actionable mtDNA mutations in iPSCs
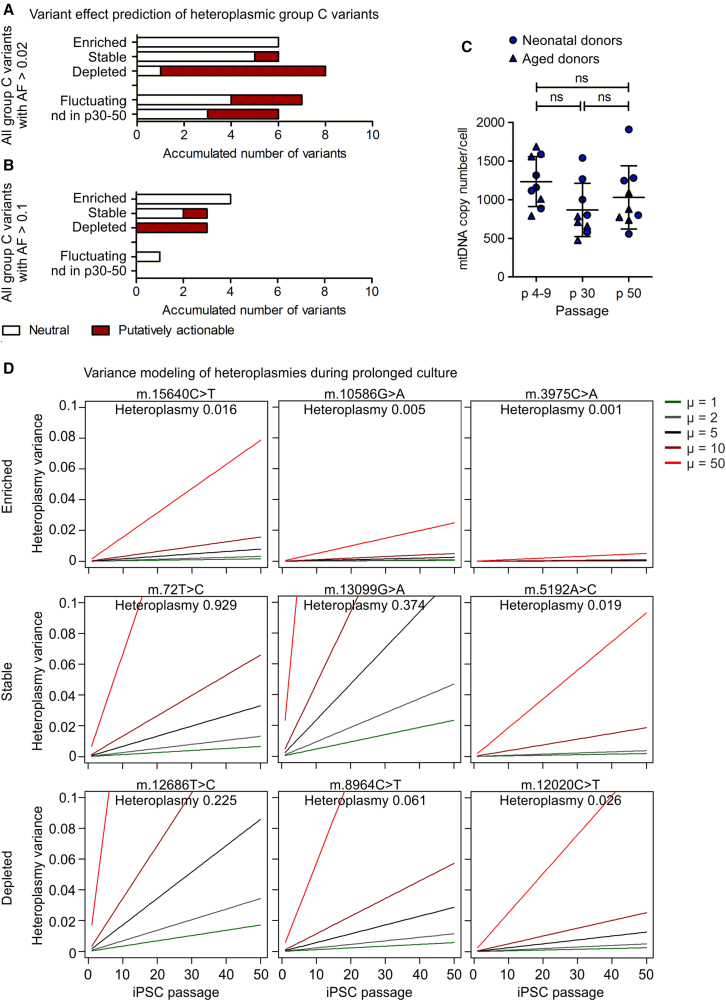
Twenty-six iPSC clones at early passage (on average p6.5) and seven of them additionally at intermediate (p30) and late passage (p50) were analyzed by mtDNA sequencing. Heteroplasmic variants (group C) were divided into the groups of enriched variants (defined as >10-fold increase in heteroplasmy level during culture), stable variants (with fold change ∼1), depleted variants (decreased by >10-fold), and fluctuating variants (change of heteroplasmy level did not allow confident classification into one of the above-mentioned groups). The group of “nd in p30-50” comprises variants that were detected in iPSC clones that were only analyzed in the early passage. The ratio of neutral and putatively actionable iPSC-enriched variants within each group was determined. Effect prediction is based on a consensus of in silico prediction algorithms (snpEff impact, CADD, Condel, and HmtVar).
(A) Analysis including all heteroplasmic group C variants with minimum AF 0.02 at any passage of iPSC expansion culture. N = 10 donors.
(B) Analysis including all group C variants with minimum AF 0.1 at any passage. N = 10 donors.
(C) Average mtDNA copy number per iPSC clone in different passages of culture expansion. N = 9 clones. One-way ANOVA with Tukey's multiple comparison test.
(D) Modeling of heteroplasmy variance over passages, exemplarily for three variants with high, intermediate, and low heteroplasmy levels for each group. The model for genetic drift was adapted from Aryaman et al. (2019) and allows prediction of variance of the heteroplasmy level and, therefore, possible change of the heteroplasmy levels over time. As a neutral model, it predicts heteroplasmy variance only based on random gene drift, while selection forces can be incorporated by an additional coefficient. For the different heteroplasmy levels in iPSCs with N = 1,000 mtDNA molecules, the heteroplasmy variances in a neutral situation (green line) and with selection acting with various strength (μ = 2, 5, 10, and 50) are shown.