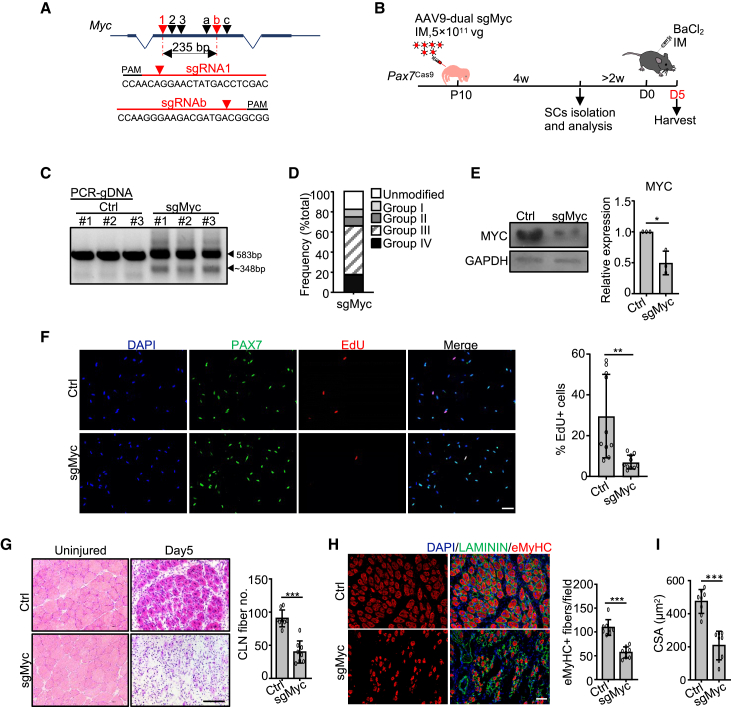
Figure 5.
CRISPR/Cas9/AAV9-sgRNA-mediated genome editing of Myc hinders SC activation and muscle regeneration
(A) Dual sgRNAs targeting Myc locus.
(B) Illustration of the experimental design.
(C) PCR to test the cleavage efficiency. Wild-type (583 bp) and cleaved (~348 bp) fragments are indicated by arrowheads.
(D) Distribution of the total sequencing reads.
(E) Left: MYC protein level. Right: the band intensity. n = 3 mice.
(F) FISCs were labeled with EdU for 24 h and the percentage of EdU+ cells was quantified. n = 10 mice (an average of 25 fields/mouse). Scale bar, 50 μm.
(G) Left: H&E staining at 5 days post injury. Right: the myofibers with CLN per field. n = 8 mice (an average of six fields/mouse). Scale bar, 100 μm.
(H) Left: immunostaining of eMyHC and LAMININ. Right: the number of eMyHC+ myofibers. n = 8 mice (an average of seven fields/mouse). Scale bar, 50 μm.
(I) The CSA of the newly formed fibers with CLN. n = 8 mice. All the bar graphs are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S5 and S6.