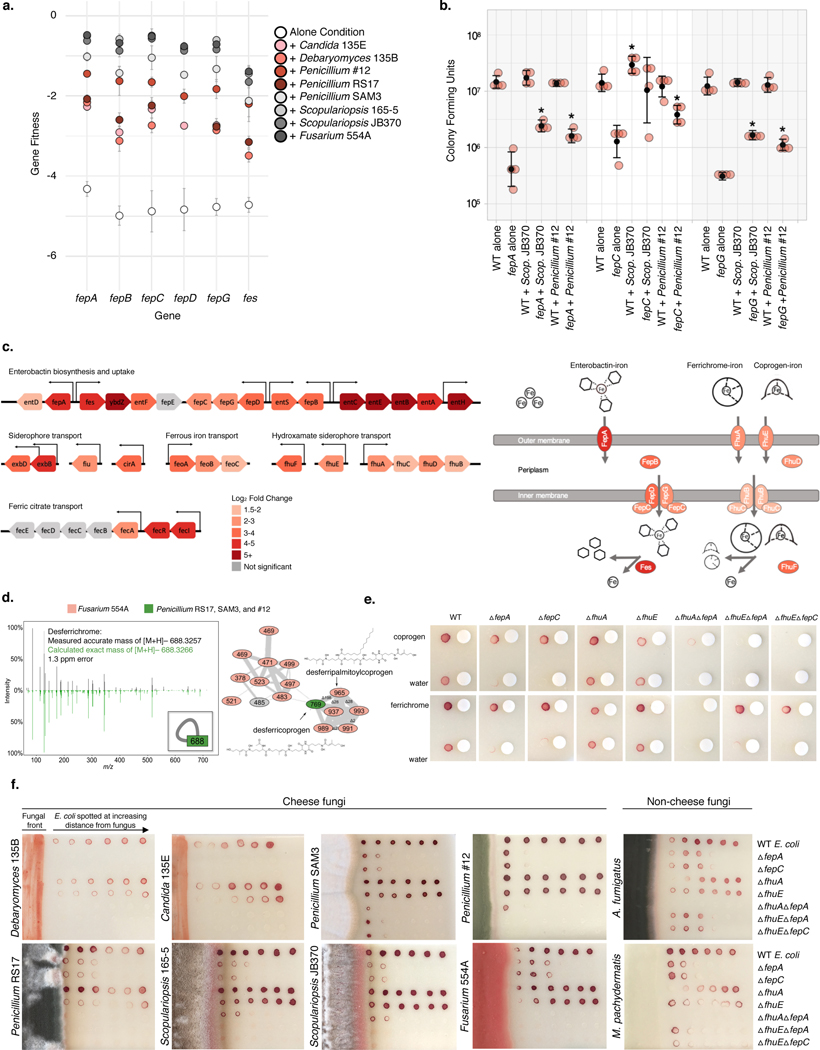
Figure 5: Utilization of fungal siderophores by E. coli.
a, RB-TnSeq fitness values for fep operon genes in alone or with-fungi conditions, showing an increase in fitness in the presence of fungal species. Fitness values are not shown for non-significant differences from alone. Error bars show standard deviation of fitness values from the mean. b, Colony forming units of WT E. coli and Δfep mutants after 7 days of 1:1 competitive growth on CCA, pH 7. Competitions between the two E. coli strains were performed either with no fungus present (“alone”) or with Penicillium sp. str. #12 or Scopulariopsis sp. str. JB370. N=4 biologically independent experiments, error bars show standard deviation from the mean (black circle). Asterisks indicate significantly different growth in the presence of a fungus relative to growth without the fungus (“alone”) based on a two-sided two-sample t-test p-value < 0.05. Exact p-values associated with asterisks (from left to right): 0.001, 0.007, 0.034, 0.022, 0.0001, 0.0006. c, E. coli iron-related genes upregulated in the presence of Penicillium sp. str. #12. Significance cutoff made at abs(log2(fold-change)) >1.5 and adjusted p-value <0.05. Differential expression analysis was performed using the default function DESeq87 which performs a Wald test with Benjamini-Hochberg75 correction for multiple comparison testing. Exact p-values are available in Supplementary Table 14. d, Fungal siderophores identified by mass spectrometry. Inset in the left box shows the node that represents the desferrichrome fragmentation pattern depicted while the network on the right represents coprogen-related molecules. Coprogen B and ferrichrome were found by matching fragmentation patterns to library spectra. Both identifications were confirmed using retention time and fragmentation matching to a purchased standard. e, Visual assays of Δfep mutant growth with purified siderophores coprogen and ferrichrome. f, Visual assays of E. coli mutant growth at varying distances from pre-cultured cheese fungi, A. fumigatus (soil, human pathogen), and M. pachydermatis (skin commensal). For e, and f, growth was performed on CCA. Tetrazolium chloride, a red indicator of cellular respiration, was added to the medium to visualize colony growth on opaque CCA.