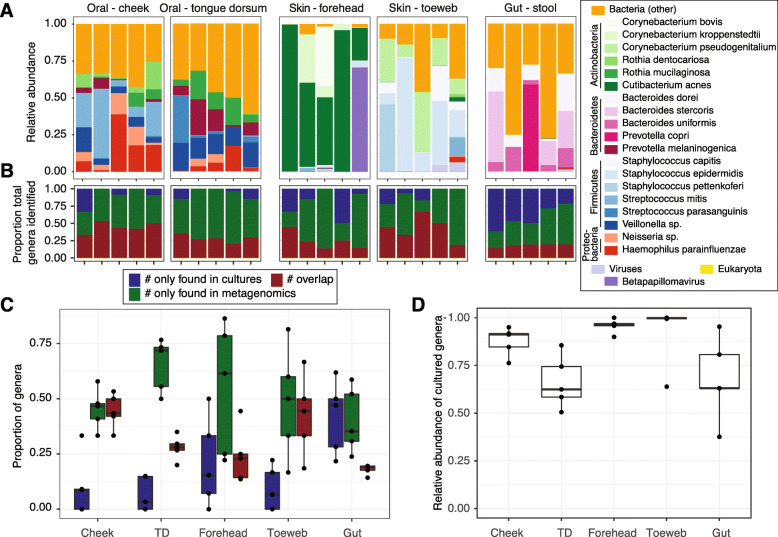
Fig. 2.
Metagenomic reconstructions of community composition and representation by cultivars. A) Relative abundance plots of oral, skin and gut samples from this study; each bar is an individual sample and the top 20 most abundant species are plotted. Lesser abundance bacteria, fungi, and viruses are collectively represented by their respective kingdom. Figure S1 visualize higher taxonomic levels. The proportion of bacterial genera cultivated or identified through sequencing for each sample, B), or across samples shown by boxplot, C). Red shows the proportion of bacterial genera identified through both cultivation and metagenomics, and blue and green show the proportion of bacterial genera identified by only one method. D) The total relative abundance of the original metagenomic sample (bacteria only) that is accounted for by the genera cultivated (overlap in C)