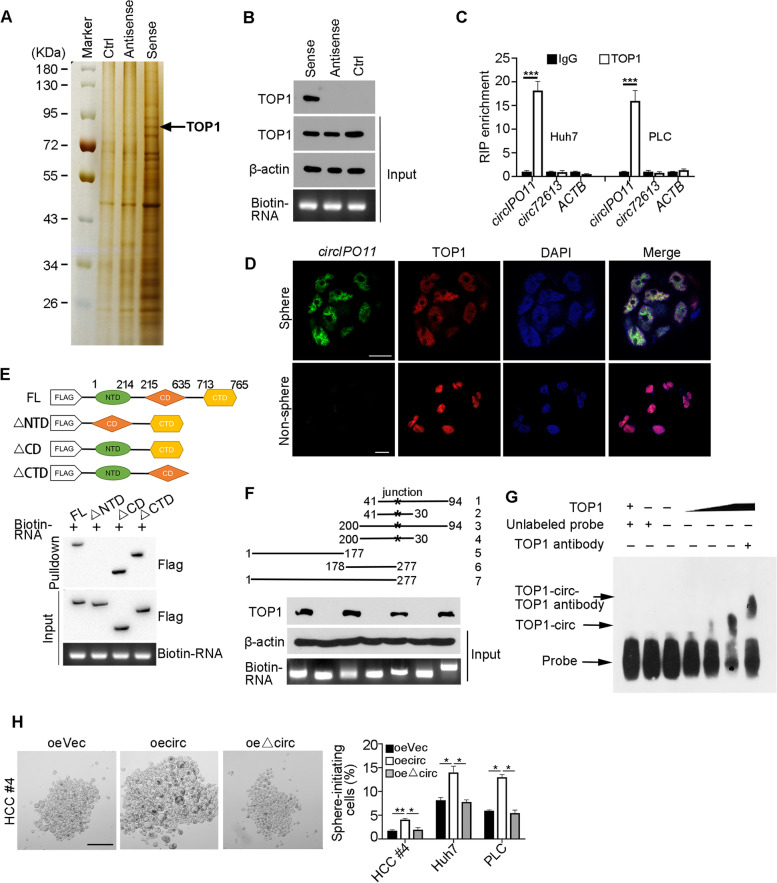
Fig. 4.
CircIPO11 associates with TOP1 in liver CSCs. A RNA pulldown assay was performed in oncosphere cell lysates using biotinylated circIPO11 junction sequences (sense), antisense and IPO11 intron sequences (Ctrl), followed by mass spectrometry. Differential band to bind circIPO11 was identified as TOP1 (black arrow). B Interaction of circIPO11 with TOP1 was confirmed from oncospheres derived from HCC samples. β-actin was used as a loading control. C Huh7 and PLC oncospheres were used for RIP assay, and followed by qRT-PCR. circRNA72613 and ACTB were used as controls. Results are shown as means ± SD. D CircIPO11 was annotated by RNA FISH, followed by immunofluorescence staining of TOP1 in oncosphere and non-sphere cells. Scale bar, 30 μm. E Domain mapping analysis of circIPO11-binding domains of TOP1 protein. Different domains of TOP1 protein were incubated with circIPO11, followed by RNA pulldown assay and Western blot. NTD, N-terminal domain; CD, core domain; CTD, C-terminal domain. F Truncated fragments of biotinylated circIPO11 were incubated with HCC oncosphere lysates, followed by RNA pulldown assay and Western blot (lower panel). Schematic diagram of circIPO11 truncated fragments (upper panel). G Biotin-labeled circIPO11 (nt 31–94) RNA probe was incubated with TOP1 for RNA EMSA. H CircIPO11 and ΔcircIPO11 were overexpressed in HCC primary cells and HCC cell lines, followed by oncosphere formation assay. Representative images (left panel) and statistical results are shown (right panel). Scale bar, 500 μm. *P < 0.05; **P < 0.01; *** P < 0.001 by two-tailed Student’s t test. Data are representative of at least three independent experiments