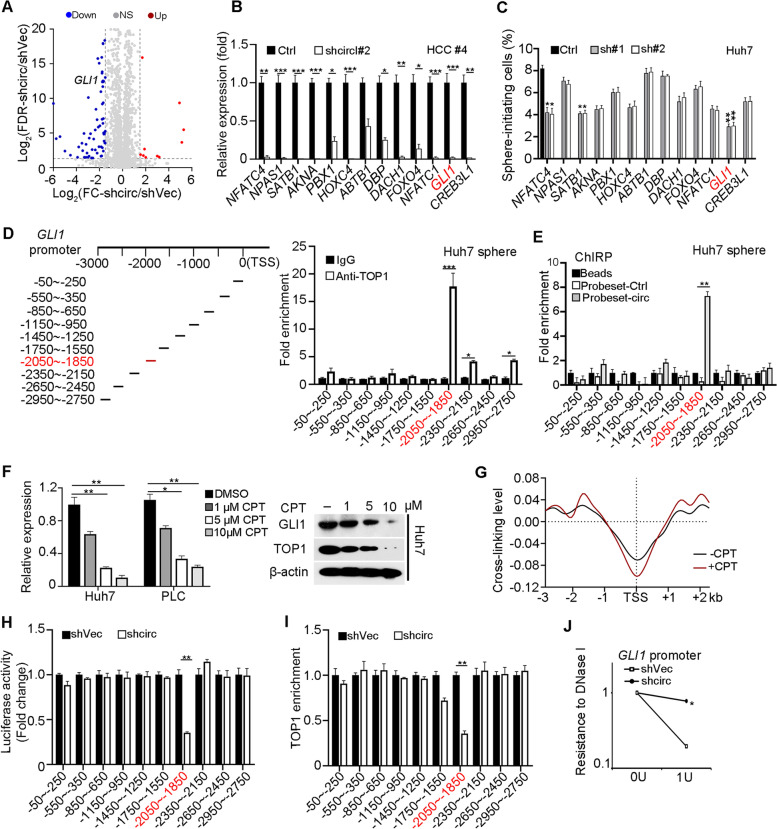
Fig. 5.
CircIPO11 recruits TOP1 onto GLI1 promoter to activate its expression. A Volcano plot of differentially expressed transcription factors in circIPO11-depleted and control liver CSC cells. B Expression levels (normalized to ACTB) of top 13 downregulated transcription factors in HCC primary cells were detected by qRT-PCR, and shVec as Ctrl for each gene. Results are shown as means ± SD. C Each of the top 13 downregulated transcription factors was silenced in HCC cell lines by shRNA, followed by oncosphere formation assay (n = 3). D ChIP was performed to identify binding regions of TOP1 on GLI1 promoter in HCC oncospheres, followed by qPCR. IgG enrichment served as a control. Results are shown as means ± SD. E CHRIP was performed to identify GLI1 promoter enrichment of circIPO11 in HCC oncospheres (n = 3). F Expression of GLI1 in HCC cells treated with different concentrations of CPT, followed by qRT-PCR and Western blot. Results are shown as means ± SD. G Psoralen photobinding assay was performed to detect superhelix structure on GLI1 promoter. H Luciferase reporter assays were performed in circIPO11-depletion and control cells. Data are shown as means ± SD. I ChIP-qPCR analysis of TOP1 enrichment on GLI1 promoter in circIPO11-depletion and control cells. Results are shown as means ± SD. J CircIPO11-depletion reduced chromatin accessibility at the GLI1 promoter by DNase I digestion assays. Results are shown as means ± SD. *P < 0.05; **P < 0.01; *** P < 0.001 by two-tailed Student’s t test. Data are representative of at least three independent experiments