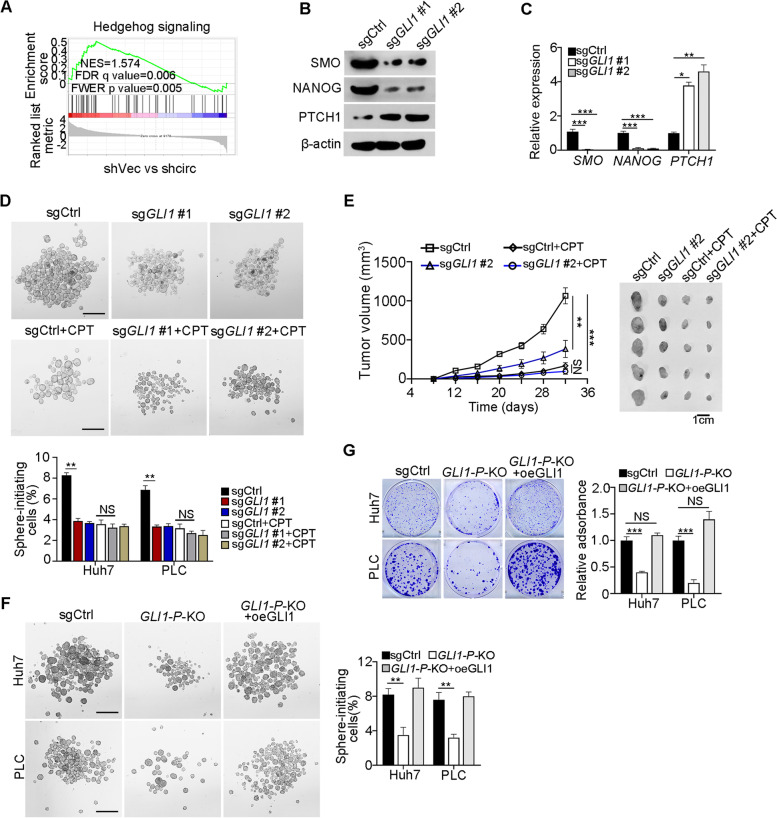
Fig. 6.
GLI1 promotes Hh signaling activation to drive liver CSC self-renewal. A GSEA indicated significantly altered genes were enriched in Hh signaling in circIPO11 depleted liver CSC cells. NES, normalized enrichment score; FDR, false discovery rate; FWER, familywise error rate. B GLI1-deleted HCC primary cells were generated by CRISPR/Cas9 approach. Whole cell lysates of GLI1 KO cells were examined with SMO, Nanog and PTCH1 antibody. C Hh signaling target genes and downstream stemness genes were tested in GLI1 KO cells by qRT-PCR. Data are shown as means ± SD. D Oncosphere formation assay was performed by GLI1-deleted HCC cell lines. Representative images (upper panel) and statistical results (lower panel) are shown. Scale bar, 500 μm (n = 3 cell cultures). E 1 × 106 GLI1-deleted cells, GLI1-deleted cells with CPT, control cells and control cells with CPT were subcutaneously injected into BALB/c nude mice. Tumor-volume curves (left panel) and representative tumor images are shown (right panel). Data are shown as means ± SD (n = 5 mice). F GLI1 overexpression rescued the reduced sphere formation capacity caused by GLI1 promoter deletion. GLI1-P-KO, the binding region of TOP1 to GLI1 promoter knockout; Results are shown as means ± SD. Scale bar, 500 μm. G GLI1 overexpression rescued the reduced clone formation capacity caused by GLI1 promoter deletion. GLI1-P-KO, the binding region of TOP1 to the GLI1 promoter knockout; Results are shown as means ± SD. **P < 0.01; *** P < 0.001 by two-tailed Student’s t test. Data are representative of at least three independent experiments