ABSTRACT
The COVID-19 pandemic has been threatening the healthcare and socioeconomic systems of entire nations. While population-based surveys to assess the distribution of SARS-CoV-2 infection have become a priority, pre-existing longitudinal studies are ideally suited to assess the determinants of COVID-19 onset and severity.The Cooperative Health Research In South Tyrol (CHRIS) study completed the baseline recruitment of 13,393 adults from the Venosta/Vinschgau rural district in 2018, collecting extensive phenotypic and biomarker data, metabolomic data, densely imputed genotype and whole-exome sequencing data.Based on CHRIS, we designed a prospective study, called CHRIS COVID-19, aimed at: 1) estimating the incidence of SARS-CoV-2 infections; 2) screening for and investigating the determinants of incident infection among CHRIS participants and their household members; 3) monitoring the immune response of infected participants prospectively.An online screening questionnaire was sent to all CHRIS participants and their household members. A random sample of 1450 participants representative of the district population was invited to assess active (nasopharyngeal swab) or past (serum antibody test) infections. We prospectively invited for complete SARS-CoV-2 testing all questionnaire completers gauged as possible cases of past infection and their household members. In positive tested individuals, antibody response is monitored quarterly for one year. Untested and negative participants receive the screening questionnaire every four weeks until gauged as possible incident cases or till the study end.Originated from a collaboration between researchers and community stakeholders, the CHRIS COVID-19 study aims at generating knowledge about the epidemiological, molecular, and genetic characterization of COVID-19 and its long-term sequelae.
KEYWORDS: COVID-19, SARS-CoV-2, Cooperative Health Research In South Tyrol (CHRIS) study, general population, novel coronavirus
Background
The COVID-19 pandemic has been threatening the healthcare systems globally, with heavy health, social, and economic consequences [1], which revealed the unpreparedness of the healthcare surveillance systems to spot, trace and contain infections, partly attributable to the peculiarities of the disease-causing virus, the SARS-CoV-2. The virus can be transmitted days before onset of symptoms [2] and most COVID-19 symptoms are not easily distinguishable from those of other respiratory infections [3]. Additionally, the disease takes advantage of conspicuous asymptomatic virus spread [4]. While much has been learned through the analysis of severe and hospitalized cases, investigating the distribution of SARS-CoV-2 infection in the general population remains a fundamental step to gain insight on the infection dynamics. This is crucial in order to provide political and healthcare authorities with realistic information to decide on adequate measures to counter the pandemic.
Furthermore, it is of primary importance to answer questions about the extent of secondary and familial transmissions, the effectiveness of precautionary directives, the duration of antibody response, and the identification of genetic, endogenous, and environmental factors that determine COVID-19 course and severity. The latter question can be best tackled by studies with genetic, epidemiological, and health information collected before the onset of the pandemic. Ideally, studies on SARS-CoV-2 infection should be nested within existing population-based studies [5,6].
In Northern Italy, SARS-CoV-2 infection prevalence was globally very high during the first pandemic phase, between late winter and early spring 2020 [3,7,8], although marked differences were observed across places, even within short physical distances. For instance, in South Tyrol, a small autonomous Italian province with about half a million inhabitants, against an overall 3.3% prevalence of individuals positive to a serum antibody test [9], valleys with a prevalence of almost 30% were also observed [3]. At the outbreak of the pandemic, in this region, the first active follow-up of the Cooperative Health Research In South Tyrol (CHRIS) study was ongoing [10]. CHRIS is a large population-based study that implements a dynamic informed consent model and had already completed the baseline assessment of 13,393 adults in 2018, with collection of accurate measurements of participants’ health, lifestyle, and individual exposures, as well as the biobanking of blood, urine, and DNA samples. A complete genetic profile of ~20 million genetic variants of each participant as well as whole-exome sequence data of a subset thereof were made available.
Following the pandemic initial burst, the follow-up phase of the CHRIS study had to be discontinued. At that time, we decided to set up a new, nested study to investigate SARS-CoV-2 infections and related COVID-19 disease: the CHRIS COVID-19 study. The main aims were to estimate the active prevalence and the cumulative incidence of SARS-CoV-2 infection in the general population of the Middle and Upper Val Venosta/Vinschgau at the end of the strict national lockdown after the first pandemic wave, and then to prospectively identify incident cases among CHRIS participants and their household members. Furthermore, the study aims at monitoring the pattern of individual antibody response over time and investigating environmental, behavioral, clinical, biological, and genetic risk factors associated with COVID-19 incidence and severity. Finally, within the framework of the ongoing CHRIS study, the CHRIS COVID-19 study will allow assessment of the long-term effects of SARS-CoV-2 infection on human health. Here, we present the design and outline the key features of the study.
Methods
Background information on the CHRIS study design and organization
The CHRIS study is a prospective study set up in 2011 in the Middle and Upper Val Venosta/Vinschgau, South Tyrol, Italy. Recruited into the studies were 13,393 participants from 13 municipalities, each one characterized by a central town, small villages, and scattered mountain farms. Settlements are located at an altitude of 600 to 2000 m above sea level. Participants cover more than one-third of the target region population. The study protocol [10] as well as details on the biological sampling and measurements [11], electrocardiographic and smoking measurements [12], and neurological questionnaires [13–15], have been previously described. At the study center, all participants underwent physical examinations, including anthropometry, 10 s and 20 min electrocardiographic analysis, blood pressure measurement, and tremor assessment. They also underwent a structured clinical interview and completed self-administered questionnaires. Ninety standard blood and urine parameters were measured [11] along with targeted and untargeted metabolomic analysis [16]. All individuals underwent genotyping and 3600 of them whole-exome sequencing. To date, high-quality imputed genotypes are available for all study participants at ~20 million genetic variants. Microbiome data in a subset have also been collected [17], thus allowing the opportunity to assess the role of microbiota in COVID-19 severity, as recently suggested [18]. Finally, an extensive biobank has been created that stores several aliquots of serum, plasma, urine, DNA, and buffy coats for all participants [10].
Aims of the CHRIS COVID-19 study
The CHRIS COVID-19 was designed to meet the following objectives:
To estimate the distribution of SARS-CoV-2 infection cases in a rural Alpine area since 1 February 2020;
To estimate the proportion of asymptomatic individuals among COVID-19 positive cases;
To characterize transmission within families;
To assess the relationship between antibody response and disease severity;
To model the evolution of antibody response over time;
To identify environmental, molecular and genetic risk factors and comorbidities associated with SARS-CoV-2 infection rate and COVID-19 incidence and severity;
To identify post-infection long-term health consequences.
Study design and participants
The CHRIS COVID-19 study is organized in three stages (Figure 1)
Stage 1: A stratified random sample of CHRIS study participants was selected to represent the adult population of the Middle and Upper Val Venosta/Vinschgau; this sample replied to an online or paper questionnaire (baseline questionnaire, Additional File 1), underwent a molecular test based on a nasopharyngeal swab and a serum antibody test, with the aim to assess the baseline situation of the region after the first pandemic wave.
Stage 2: All 13,393 CHRIS study participants and their consenting cohabitants were submitted an online questionnaire (baseline questionnaire, Additional File 1) to report past and current health status, with the emphasis on potential SARS-CoV-2 infection. A shorter version of the questionnaire (follow-up questionnaire, Additional File 2) has been submitted repeatedly to participants every 4 weeks for an update of their health status relative to the ongoing COVID-19 situation. All individuals gauged at risk of positivity to SARS-CoV-2 infection and their cohabitants have been invited for a nasopharyngeal swab molecular test and a serum antibody test at the study center.
Stage 3: To trace and monitor antibody response over time, all individuals testing positive to either the nasopharyngeal or the serum test in Stages 1 or 2 have been invited to repeat the serum antibody test every three months for a year, since their first measurement.
Figure 1.
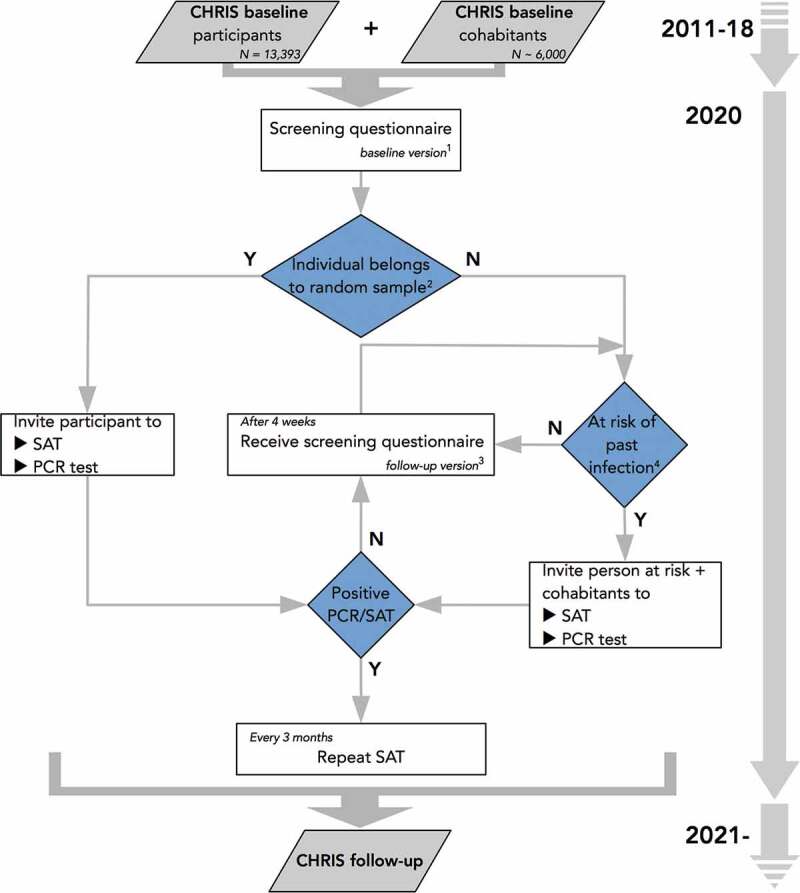
Flowchart of the CHRIS COVID-19 study. The study is based on all 13,393 participants to the CHRIS study (2011–18) and their cohabitants. Abbreviations: SAT, serum antibody test; PCR, polymerase chain reaction. Notes: 1The baseline questionnaire is reported in Additional File 1; 2A random sample of 1450 (augmented to 1812) has been extracted from the 13,393 CHRIS participants to derive population-representative estimates of SARS-CoV-2 infection in the study area (see Methods); 3The follow-up questionnaire is reported in Additional File 2; 4Risk of past infection is derived based on responses to the self-administered questionnaires.
Details of each stage and the respective operations are given below.
Stage 1 – baseline prevalence estimation
This stage was designed to define the baseline situation in our study, that is, to estimate the cumulative incidence of SARS-CoV-2 infections in the valley between February 1 and 28 August 2020. We estimated that a sample size of 1450 individuals would suffice to estimate a cumulative incidence between 0.01% and 1.1% with a confidence level of 99.0%, or a cumulative incidence of ≤0.5% with confidence level of 98.2% (Table 1). Estimates were based on the Clopper–Pearson exact method for extreme proportions [19] and obtained using the software PASS v20.0.1. The hypothesis of such low incidence values was based on the very low count of 16 COVID-19 positive cases identified by the local Healthcare System until June 2020, at which time the study protocol was submitted for ethical approval, out of >36,000 inhabitants in the study area.
Table 1.
Precision of the cumulative incidence estimate assuming a sample size of 1450 participants, by three levels of magnitude of the cumulative incidence hypothesized. Reported are the positive predictive values for the Roche Elecsys Anti-SARS-CoV-2 serum antibody test according to 100% sensitivity and 99.8% specificity declared by the manufacturer
| Hypothesized cumulative incidence | Confidence interval of the estimated cumulative incidence | Confidence level | Positive predictive value of the test (confidence interval) |
|---|---|---|---|
| 0.440% | 0.1%, 1.1% | 99.0% | 70% (33%, 85%) |
| 0.088% | 0.0%, 0.5% | 98.2% | 31% (0%, 72%) |
| 0.066% | 0.0%, 0.5% | 98.9% | 25% (0%, 72%) |
Assuming an 80% participation rate, we oversampled 1812 randomly selected individuals from all CHRIS study participants. The sample was designed to be representative of the general adult population of the study area in terms of age and sex distribution. Participants were invited to complete a screening questionnaire and to undergo both a serological antibody test and a nasopharyngeal test to identify possible SARS-CoV-2 infection. To ease participation, three recruitment centers were prompted in the lower (July 13–16), upper (July 20–22), and central locations (July 28-August 28) of the rather extensive valley.
Stage 2 – prospective screening
Between 13 July 2020 and 31 July 2021, all 13,393 CHRIS study participants, including those involved in the Stage 1, received an online screening questionnaire (baseline questionnaire) addressed also to their cohabitants, including minors, house attendants and caregivers, resulting in an estimated number of ~19,000 invited individuals. A first contact was established via postal letter, e-mail or phone call. CHRIS participants received a link along with unique credentials to an online registration portal, and were invited to register themselves and their cohabitants using separate credentials for each member. One member would act as the contact person, identified by personal tax number and verification of identity via upload of an official identification document.
The baseline questionnaire (Additional File 1) asked details about diagnosis, symptoms, and exposure to the virus, from 1 February 2020, until the day of participation. The questionnaire covered the following aspects: symptoms; comorbidities; regular therapies; previous relevant health issues; socio-demographic information; close contact with symptomatic or confirmed positive for COVID-19 persons; essential lifestyle; and vaccination. The questionnaire of the main respondent contained a section about the dwelling, while some questions were omitted in the versions for <14 year old children.
A simplified form of the same questionnaire has been sent to participants every four weeks to monitor incident events (follow-up questionnaire; Additional File 2).
According to set criteria, based on responses to the questionnaire, participants who may have had past infection have been prospectively invited along with their cohabitants (regardless of the cohabitants’ questionnaire responses) to undergo a serological test and a nasopharyngeal swab at the study center. This strategy would ensure the identification of the largest possible number of positive cases as it also would enable assessment of pre- and asymptomatic cohabitants. This approach would also allow observation of the spread of SARS-CoV-2 infections among cohabitants, for potential discrimination between infected and resistant individuals.
Stage 3 – antibody response monitoring
To evaluate the antibody response over time, individuals who tested positive to one of the tests performed at the study center (either the swab retro-transcriptase real-time polymerase chain reaction (RT-PCR) test or the serum antibody test) would be invited to repeat the serological antibody test at 3, 6, 9, and 12 months after the initial visit.
Blood sampling, serum antibody test, and nasopharyngeal swab
All individuals to be tested have been invited to undergo a nasopharyngeal swab to assess active SARS-CoV-2 infection through a RT-PCR test.
Participants in Stage 1 and Stage 2 in-person examinations had 7 ml of venous blood drawn, filling two 3.5 ml serum tubes (VACUETTE TUBE 3.5 ml Z Serum Separator Clot Activator 16 × 100 red cap-yellow ring, non-ridged), one for serum antibody testing (see below) and the other to generate four 250 µl serum aliquots for cryopreservation. Participants with no or low quantities of genomic DNA stored already in the CHRIS biobank were asked to draw three additional ml in a whole-blood EDTA tube (VACUETTE TUBE 3 ml K2E K2EDTA 16 × 100 lavender cap-black ring, non-ridged). Participants in the Stage 3 examinations have had 7 ml venous blood drawn, filling two 3.5 ml serum tubes each time, which have been used for serum antibody testing and to generate two 500 µl serum aliquots for cryopreservation.
IgG antibodies against SARS-CoV-2 have been assessed using the Roche Elecsys® Anti-SARS-CoV-2 assay based on high throughput ELISA technology, which uses a recombinant protein that represents the nucleocapsid (N) antigen. Per manufacturer declaration, the test has 99.5% sensitivity at ≥14 days post RT-PCR confirmation and 99.8% specificity. Material not used for testing will have been destroyed.
For participants in the Stage 2 examinations, an additional collection of two 20 µl whole blood microsamples (Mitra by Neoteryx) has been proposed. Participants to Stage 3 who had accepted to undergo the additional microsampling collection, have been invited to perform autonomous microsampling at home, again two microsamples of 20 µl blood each, at 1 and 2 months after the visit. Microsamples have been collected in the biobank and used for testing antibody response to infection, thus complementing the serum collection and tests.
Biobanking
Serum and whole-blood EDTA tubes have been stored at 4°C in a refrigerator until shipment to the Eurac Research Biobank at the Bolzano/Bozen hospital. At the biobank, DNA has been extracted from the 3 ml whole-blood EDTA tube using an automatic extractor (chemagic 360, Perkin Elmer). The 3.5 ml serum tube has been split into four 250 µl aliquots (Stages 1 and 2) or two 500 µl aliquots (Stage 3), which have been then frozen by direct liquid nitrogen immersion and stored at −80°C for subsequent biochemical investigations. The aliquoting has been performed using a robotic system (Starlet, Hamilton). Biological specimens have been stored using the Thermo Scientific™ Nunc™ Biobanking system. Daughter aliquots have been given unique codes that allow tracing back to the mother tubes and then to the participants codes, using the biobank management software. Microsamples collected at Stage 2 and 3 have been stored at −80°C.
Neutralizing antibody assessment
The ability of the serum to inhibit the transduction of a lentiviral vector pseudotyped with the SARS-CoV-2 spike protein will have been evaluated. As to the procedure workflow, pseudotyped vectors, transducing a gene encoding a fluorescent protein, are incubated along with scalar dilutions of serum that is then inoculated onto Huh-7 cells. Percentage of transduced fluorescent cells is next quantified using the High Content Molecular Device Image Xpress® Micro Confocal upon nuclei counterstaining with Hoechst 33,342 and the serum dilution associated with 50% inhibition of transduction (ID50 value) is finally estimated from each derived sigmoidal curve.
Outcomes
The study will initially focus on two main related outcomes: (1) detection of SARS-CoV-2 active or past infection (proxy for COVID-19); (2) severity of COVID-19.
(1) Detection of SARS-CoV-2 active or past infection will be determined by the nasopharyngeal swab RT-PCR test and the serum antibody test; notice of self-reported diagnosis of COVID-19, previous serum antibody or nasopharyngeal-based test diagnosis, will be assessed. All participants not fulfilling criteria for a possible or suspected case will be classified as negatives, therefore free of COVID-19. Sensitivity analyses of the screening questionnaire are foreseen to assess the validity of case classification (e.g. presence of featured symptoms in the absence of a positive diagnosis).
(2) Severity of COVID-19 will be defined based on the best available information. At minimum, proxy of severity will be defined based on the information obtained from the screening questionnaire. Specifically, we will refer to any reported symptoms (Additional File 1, question 2.6), symptoms duration (question 2.8), outlook for medical assistance (question 2.9), symptoms related limitations of daily activities (question 2.10), and hospitalization because of suspected or confirmed SARS-CoV-2 infection (question 2.3). In the case of a sufficient number of participants reporting hospitalization, informed consent allows us to access participants’ clinical records and classify severity based on the World Health Organization criteria [32], through integration with responses to the screening questionnaire. Additional medical record-based classifications [20] will be explored [21].
Statistical analyses
Prevalence or cumulative incidence will be estimated according to the stratified random survey design for best accuracy and precision. We will also adopt sampling weights as appropriate to account for self-selection of participants, due to non-response. For better coverage of the relatively low incidence expected at baseline, we will use the exact method for confidence interval figures, consistently with the methods used for the sample size calculation. The association between determinants and presence/absence of SARS-CoV-2 infection will be assessed using generalized linear mixed models, in order to take into account the presence of stratification factors or clustered structures of the CHRIS study data, such as households, residence, and relatedness [11]. In particular, logistic regression models will be used to analyze binary endpoints. Categorical endpoints will be fitted with a multinomial model. Quantitative endpoints will be analyzed using linear models. Incident events will be modeled using mixed Cox regression models for non-independent observations [22]. Mixed regression models for longitudinal data will be used for analysis of persistence of immunity over time (baseline + up to 4 repeated measures).
Ethical, legal, and social issues
Conducting research in an emergency requires exceptional means to organize and conduct ethically and legally sound studies. Prospective participants have been informed about the study through a massive and coordinated information strategy, which included press releases in the local newspapers, interviews with the study coordinator, presentation of the study to the public and the local authorities and stakeholders, and widespread distribution of descriptive flyers through the local pharmacies. The CHRIS study website was timely updated with the description of the study and all the necessary information (https://de.chris.eurac.edu/_startpage/chris-covid-19-studie/).
Invitation proceeded through electronic and ordinary mail. Participants were informed about the aims and perspectives of the research and signed an informed consent with detailed information about the study in order to take part in the research (Additional File 3). Participants were required to express their informed consent prior to enrollment in different ways, depending on the stage of the study and on their condition: (a) to participate in the online surveys, participants had to fill in the online informed consent and upload an identification document: (b) to participate in the in-person testing visits, participants were asked to fill in and sign a specific paper-based informed consent; (c) in the presence of minors, parental authority have been in charge of consenting and submitting the questionnaire or to allow participation to the in-person testing stages.
Access to COVID-19 related data in the medical records through controlled access by the Healthcare System only is foreseen. Participants had the option to give consent for their clinical data, relative to COVID-19 and related diseases, to be transmitted in encrypted form to Eurac Research to conduct scientific research. Any report of clinically relevant results to study participants has been handled directly by the Healthcare System.
The CHRIS COVID-19 study complies with European legislation (EU GDPR 2016) and the Italian law on personal data protection where it differs from the European legislation, the Declaration of Helsinki (as amended), the World Medical Association Declaration of Taipei 2016 on ethical considerations regarding health databases and biobanks, the Convention on Human Rights and Biomedicine (Oviedo 1997 as amended), and the Italian law on scientific research. It follows the international guidelines of the Council for International Organizations of Medical Sciences, the National Bioethics Committee guidelines and the National Committee for Biotechnology, Biosafety and Life Sciences guidelines on the collection of biological samples for research purposes (2009).
The CHRIS COVID-19 study was authorized by the Ethics Committee of the Healthcare System of the Autonomous Province of Bolzano/Bozen with deliberation number 53–2020 of May 27 and 22 July 2020.
Discussion
After almost 20 years of presence in the Val Venosta/Vinschgau district with the MICROS [23] and CHRIS population-based studies, and the close relationship built with local communities, the CHRIS COVID-19 study was born as a response to a bottom-up approach that reached the researchers from the citizen representatives’ initiative. The incoming health emergency and the awareness of the establishment of a longitudinal study in the area motivated the population to seize the opportunity to contribute to the advancement of scientific knowledge for the good of the population’s health. The CHRIS COVID-19 study therefore consists of two complementary components: a branch that aims to quantify the presence of infections in the valley during the pandemic, and a branch that exploits the existing database from the previous CHRIS study in order to understand which individual factors might be associated with a greater susceptibility to both infection and COVID-19 severity. The creation of a cryopreserved genomic and serum biobank will be an important resource to answer future new scientific questions.
Being nested in the ongoing CHRIS study, the CHRIS COVID-19 study will allow assessment of the long-term effects of SARS-CoV-2 infection. The recently defined post-acute COVID-19 syndrome includes symptoms lasting for more than 12 weeks since COVID-19 onset, not explained by other diseases, and affecting multiple organs with variable severity [24]. Onset of hematologic, cardiovascular, neuropsychiatric, renal and endocrine sequelae could be monitored among CHRIS participants, given data collected before SARS-CoV-2 emergency.
The CHRIS COVID-19 study presents novelties and peculiarities compared to its parent CHRIS study. For the first time, a re-consent was submitted to participants entirely online (i.e.: the consent for the survey). This was made possible by the consolidated experience of the CHRIS study with dynamic consent, which is used in the CHRIS study since 2011 [10]. Dynamic consent is a consent model that allows participants to modify their participation options over time, while being regularly informed about the study and research developments [25,26]. As it largely relies on information technology, the dynamic consent becomes more and more relevant in situations of enhanced digital technology use, such as in the case of a pandemic, when human mobility is limited. The capillary and strong information strategy laid the foundation for a strong participation under extraordinary circumstances. Another novel aspect is that, for the first time, family members, non-CHRIS cohabitants (who are not necessarily relatives) and minors have been invited: such an expansion of the CHRIS study is being allowed by the adaptability of the dynamic consent in responding to research needs.
The study has multiple public health prospects. First, estimating COVID-19 prevalence from a putative representative cohort will give a reasonably unbiased snapshot of the spread of the virus in the valley and will inform future strategies for such endeavor to avoid limitations and act timely. Furthermore, active tracking of incident cases will help modeling the infection dynamics, and evaluating health surveillance and prevention policies. CHRIS COVID-19 is also in the position to deepen important research questions. For instance, it will be possible to model the patterns of infection within households, investigating factors of either resilience or susceptibility to infection and its complications.
Families of both infected and uninfected members could be explored using available exome sequencing data, searching for variants responsible of natural resistance to the infection. Similarly to HIV natural resistance due to Δ32 deletion in CCR5 or variants in DPP4 decreasing infection potential of MERS-CoV [27], we cannot exclude that variants in ACE2 or other genes, essential for SARS-CoV-2 entry in cells, could fully prevent the infection. Currently, the investigation of SARS-CoV-2 genetic natural resistance remains unexplored.
The effective enrollment strategy can be exploited to tackle open scientific questions. In particular, the involvement of families may provide tentative answers to one of the most controversial issues in SARS-CoV-2 research, namely the contribution of minors to the spread of the virus. A recently published systematic review and meta-analysis showed that overall children do contribute to a small extent to transmission clusters, and that they are less susceptible to secondary attack rate, compared to adults [28]. CHRIS COVID-19 has the potential to shed more light on this topic.
Monitoring positive cases over time will allow us to assess whether the duration of the antibody response triggered by SARS-CoV-2 infection is resilient over time, and what are the factors associated with variability in circulating antibody titers. While antibody persistence exhibits considerable heterogeneity in different studies [29], recent evidence suggests that antibodies can persist up to eight months since initial infection [30]. Amidst the first massive vaccination campaign, monitoring participants over time will also allow assessment of their response to vaccination, which was introduced as an extra item of investigation in the questionnaire since the start of the campaign. First evidence shows that the mRNA vaccine triggered a stronger and quicker immune response in individuals who recovered from COVID-19 (seropositive), compared to seronegative. Final antibody levels were similar across groups, with slightly higher levels in seropositive individuals [31]. The CHRIS COVID-19 study is therefore suitable to address questions around anti-SARS-CoV-2 vaccine efficacy and antibody response in real-world scenarios.
The presence of another COVID-19 population-based study in South Tyrol [3] with comparable study design may favor a ‘discovery-replication’ approach, where one study can be used to generate hypotheses and the other study can be used to replicate or refute the generated hypothesis. Further, one sample may support building predictive models, which can then be tested or validated in the other sample. The common cultural background among these two target populations is an advantage to this framework.
The identification of genetic factors in humans that may influence COVID-19 susceptibility, severity, and outcomes may lead to an acceleration in the development of solutions to counter COVID-19. Likewise, it would allow for stratification of the infected persons into higher risk groups that could be offered earlier access to mitigating therapies. The identification of genes and genetic variants associated with clinical-epidemiological outcomes may be crucial in several ways: drug repurposing, identification of people with higher risk or, conversely, people who seem to be naturally protected, and otherwise contribute to the increase of general biological knowledge on SARS-CoV-2 and proximal infections and diseases. The data obtained from the study will be used to perform genome-wide association scans to identify genetic variants associated with both the risk and severity of infection leading to COVID 19. In this context, CHRIS COVID-19 is already a partner of the COVID-19 Host Genetics Initiative (www.covid19hg.org), which has been conducting genome-wide association studies on several millions participants globally [32].
In conclusion, born from a collaboration between researchers and communities, the CHRIS COVID-19 study will answer the short-term public health needs to quantify SARS-CoV-2 spread, while generating new knowledge about the epidemiological, molecular, and genetic characterization of COVID-19.
Supplementary Material
Acknowledgments
At Eurac Research, we thank the study team (Roselinde Gunsch, Karin Bystrianska, Brunhilde M. Grasser, Benedikta Linter, Liane Parth, Renate Telser, and Elena Cannavò), the biobank team (Giulia Caprioli, Ilaria Bozzolan, Flavio Matassoni), and information technology team (Daniele Di Domizio, Johannes Martin, Yuri D’Elia), and several colleagues who contributed to the implementation of the project with literature review, contribution of ideas, curation of the legal aspects, and field work (Cristina Berloffa, Hagen Blankenburg, Annalisa Boscaro, Chiara Cantaloni, Corrado Corti, Fabiola Del Greco M, Cosimo Delli Carpini, Francesca Di Leva, Laura Eccel, Selene Fornaca, Beatrice Giaier, Eva König, Irene Lunger, Ilaria Pagano, Massimiliano Pellegrini, Anne Picard, Athina Raftopoulou, Johannes Rainer, Diana Anna Riekschnitz, Marcelo Damian Rosato Siri, Maja Schlittler, Katharina Tschigg, Claudia Volpato, Vladimir Vukovic), as well as the communication team that supported the dissemination campaign for the study (Elena Munari and Laura Defranceschi). We thank Dr. Martin Matscher, Sonja Gorfer, and Isolde Maria Blaas from the Healthcare System of the Autonomous Province of Bolzano/Bozen, and Tanja Plörer (Municipal police of Laces/Latsch; Val Venosta/Vinschgau District Community; and Val Venosta/Vinschgau White Cross) for the organizational support. The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the Open Access publication costs.
Funding Statement
The CHRIS COVID-19 study is supported by Eurac Research, the South Tyrolean Health Authority, and the Department of Innovation, Research and University of the Autonomous Province of Bolzano. The support is meant to be both financial and through in-kind contributions, such as the provision of human and instrumental resources as well as organizational and logistical help. The present research was conducted within the project ‘PACE: Partnership to Accelerate Covid-19 rEsearch in South Tyrol’, funded by Department of Innovation, Research and University of the Autonomous Province of Bolzano within the 2019–2021 Research Program (unique project code: D52F20000770003).
List of abbreviations
If abbreviations are used in the text they should be defined in the text at first use, and a list of abbreviations should be provided.
ACE2: Angiotensin-converting enzyme 2
ASTAT: Provincial Institute of Statistics
CCR5: C-C chemokine receptor type 5
CHRIS: Cooperative Health Research In South Tyrol
COVID-19: Coronavirus Disease 2019
DNA: Deoxyribonucleic acid
DPP4: Dipeptidyl peptidase 4
GDPR: General Data Protection Regulation
GFP: Green Fluorescent Protein
HIV: Human Immunodeficiency Viruses
ISTAT: Italian National Institute of Statistics
MERS-CoV: Middle East Respiratory Syndrome – Coronavirus
mRNA: messenger Ribonucleic Acid
RT-PCR: retro-transcriptase real-time polymerase chain reaction
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2
Authors’ contributions
Drafted the manuscript: CP, GB, LF, CXW, RB, FSD, MG, RM
Study design: CP, FSD, MG, RM, PPP
Definition of the ethical framework: DM
Biobank and laboratory operations and measurements: ADG, MP
Intellectual contribution to the study design: CF, JR, AAH, MM, AM, SL, HM, HW, RR, CD, MP, GW, EL, AC
Study implementation: MG, CE, VCA
Critical revision of the manuscript: All authors
All authors have read and approved the manuscript
Availability of data and materials
All questionnaires, information, and consent forms are provided in the Additional Files 1, 2, and 3. They can be used for scientific research purpose. Commercial use is not authorized. The study principal investigators are available to share information about the standard operative procedures that might be useful to other researchers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Consent for publication
All Authors have read and approved the manuscript and approve its publication.
Ethics approval and consent to participate
The CHRIS COVID-19 study was approved by the Ethics Committee of the Healthcare System of the Autonomous Province of Bolzano/Bozen with deliberation number 53-2020 of May 27 and 22 July 2020. All participants provide online-based or written informed consent upon enrollment, as specified in the methods.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wei WE, Li Z, Chiew CJ, et al. Presymptomatic transmission of SARS-CoV-2 - Singapore, January 23-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Melotti R, Scaggiante F, Falciani M, et al. Prevalence and determinants of serum antibodies to SARS-CoV-2 in the general population of the Gardena Valley. Epidemiol Infect. 2021;149:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lavezzo E, Franchin E, Ciavarella C, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584(7821):425–429. [DOI] [PubMed] [Google Scholar]
- [5].Ho FK, Celis-Morales CA, Gray SR, et al. Modifiable and non-modifiable risk factors for COVID-19, and comparison to risk factors for influenza and pneumonia: results from a UK Biobank prospective cohort study. BMJ Open. 2020;10(11):e040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Intyre KM, Lanting P, Deelen P, et al. Lifelines COVID-19 cohort: investigating COVID-19 infection and its health and societal impacts in a Dutch population-based cohort. BMJ Open. 2021;11:e044474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pagani G, Conti F, Giacomelli A, et al. Seroprevalence of SARS-CoV-2 significantly varies with age: preliminary results from a mass population screening. J Infect. 2020;81(6):e10–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stefanelli P, Bella A, Fedele G, et al. Prevalence of SARS-CoV-2 IgG antibodies in an area of northeastern Italy with a high incidence of COVID-19 cases: a population-based study. Clin Microbiol Infect. 2021;27(4):633.e1–633.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].ISTAT . Primi risultati dell’indagine di sieroprevalenza SARS-CoV-2. ISTAT, Italian National Statistical Institute, 2020 Aug 3. http://www.salute.gov.it/imgs/C_17_notizie_4998_0_file.pdf
- [10].Pattaro C, Gögele M, Mascalzoni D, et al. The Cooperative Health Research In South Tyrol (CHRIS) study: rationale, objectives, and preliminary results. J Transl Med. 2015;13:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Noce D, Gögele M, Schwienbacher C, et al. Sequential recruitment of study participants may inflate genetic heritability estimates. Hum Genet. 2017;136(6):743–757. [DOI] [PubMed] [Google Scholar]
- [12].Murgia F, Melotti R, Foco L, et al. Effects of smoking status, history and intensity on heart rate variability in the general population: the CHRIS study. PLoS One. 2019;14(4):e0215053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zanigni S, Giannini G, Melotti R, et al. Association between restless legs syndrome and migraine: a population-based study. Eur J Neurol. 2014;21(9):1205–1210. [DOI] [PubMed] [Google Scholar]
- [14].Giannini G, Zanigni S, Melotti R, et al. Association between restless legs syndrome and hypertension: a preliminary population-based study in South Tyrol, Italy. Eur J Neurol. 2014;21(1):72–78. [DOI] [PubMed] [Google Scholar]
- [15].Melotti R, Ruscheweyh R, Pramstaller PP, et al. Structural consistency of the pain sensitivity questionnaire in the Cooperative Health Research In South Tyrol (CHRIS) population-based study. J Pain. 2018;19(12):1424–1434. [DOI] [PubMed] [Google Scholar]
- [16].Paglia G, Del Greco FM, Sigurdsson BB, et al. Influence of collection tubes during quantitative targeted metabolomics studies in human blood samples. Clin Chim Acta. 2018;486:320–328. [DOI] [PubMed] [Google Scholar]
- [17].Motta BM, Grander C, Gögele M, et al. Microbiota, type 2 diabetes and non-alcoholic fatty liver disease: protocol of an observational study. J Transl Med. 2019;17(1):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yeoh YK, Zuo T, Lui GC-Y, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17(8):857–872. [DOI] [PubMed] [Google Scholar]
- [20].Murray MF, Kenny EE, Ritchie MD, et al. COVID-19 outcomes and the human genome. Genet Med. 2020;22(7):1175–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. [DOI] [PubMed] [Google Scholar]
- [22].Therneau T. Mixed effects cox models. Mayo Clinic. 2020 Jan 13. Available at: https://cran.r-project.org/web/packages/coxme/vignettes/coxme.pdf
- [23].Pattaro C, Marroni F, Riegler A, et al. The genetic study of three population microisolates in South Tyrol (MICROS): study design and epidemiological perspectives. BMC Med Genet. 2007;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nalbandian A, Sehgal K, Gupta A, et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27(4):601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Budin-Ljøsne I, Teare HJA, Kaye J, et al. Dynamic consent: a potential solution to some of the challenges of modern biomedical research. BMC Med Ethics. 2017;18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kaye J, Whitley EA, Lund D, et al. Dynamic consent: a patient interface for twenty-first century research networks. Eur J Hum Genet. 2015;23(2):141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Devaux CA, Rolain J-M, Raoult D, et al. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhu Y, Bloxham CJ, Hulme KD, et al. A meta-analysis on the role of children in SARS-CoV-2 in household transmission clusters. Clin Infect Dis. 2020; 72(12):e1146-e1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science [Internet]. American Association for the Advancement of Science, 2021. [cited 2021 Apr 22];371. Available from: https://science.sciencemag.org/content/371/6529/eabf4063 [DOI] [PMC free article] [PubMed]
- [31].Krammer F, Srivastava K, Alshammary H, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;384(14):1372–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].The COVID-19 Host Genetics Initiative . The COVID-19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28(6):715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All questionnaires, information, and consent forms are provided in the Additional Files 1, 2, and 3. They can be used for scientific research purpose. Commercial use is not authorized. The study principal investigators are available to share information about the standard operative procedures that might be useful to other researchers.


