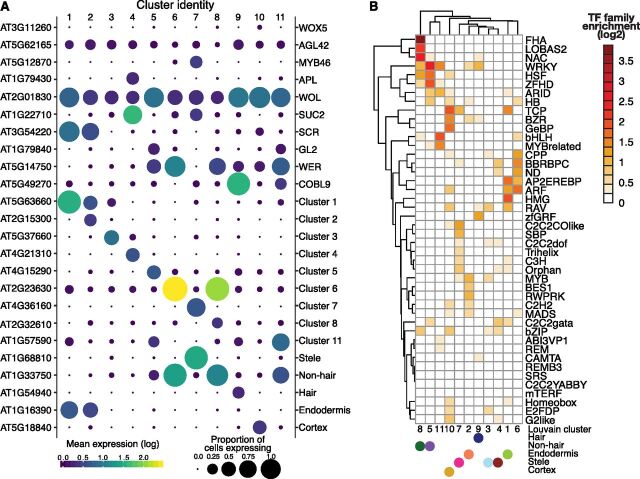
Figure 2.
Novel Cluster-Specific and Tissue-Specific Genes and Enriched Transcription Factor Motifs.
(A) The proportion of cells (circle size) and the mean expression (circle color) of genes with cluster-specific and tissue-specific expression are shown, beginning with known marker genes labeled with their common name (right) and their systematic name (left). For novel genes, the top significant cluster-specific genes are shown, followed by the top significant tissue-specific genes; both were identified by principal graph tests (Moran’s I) as implemented in Monocle 3. Note the correspondence between Louvain components and cell and tissue types. For all novel cluster-specific and tissue-specific genes, see Supplemental Table 3.
(B) Enrichments of known transcription factor motifs (O’Malley et al., 2016) 500 bp upstream of genes with cluster-specific expression compared with genome background. Motifs are specific to transcription factor gene families rather than individual genes. The plot is clustered based on similarity in enrichments with Louvain components and the cell and tissue types (solid circles) indicated.