FIGURE 4.
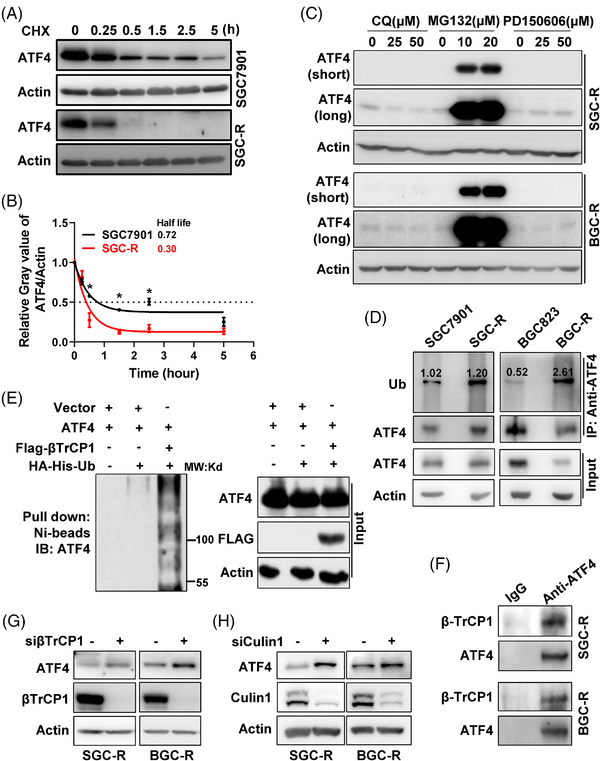
βTrCP‐enhanced ubiquitination‐dependent proteasomal degradation of ATF4 in chemoresistant cells. (A) Turnover of endogenous ATF4 in SGC7901 or SGC‐R cells under protein synthesis inhibitor cycloheximide (CHX, 50 μg/ml) incubation was detected by western blotting using anti‐ATF4 antibody. (B) The relative grey value of ATF4 compared to Actin in ‘A’ was analysed by Image J, which was further normalised to the ‘0’ time point sample. The fitted curves were drawn with GraphPad software and the half‐life of ATF4 turnover was analysed and shown. (C) Expression of ATF4 in SGC‐R (up) or BGC‐R (down) with different protein degradation inhibitors (lysosome inhibitor: CQ, proteasome inhibitor: MG132 and calpain inhibitor: PD150606) treatment was determined by western blotting. The short‐time and long‐time exposure results were shown as ATF4 (short) and ATF4 (long), respectively. (D) After pre‐treated with MG132, ubiquitination of endogenous ATF4 in sensitive or resistant cells was detected by anti‐ATF4 immunoprecipitation (IP) and anti‐Ub immunoblot. ATF4 ubiquitination was normalised to immunoprecipitated ATF4, and the normalised ratio was shown. (E) βTrCP1‐mediated ATF4 ubiquitination was determined by an in vitro ubiquitination assay in HEK293T cells. (F) The interaction of ATF4 with βTrCP1 in SGC‐R (up) or BGC‐R (down) cells pre‐treated with MG132 was analysed by anti‐ATF4 co‐immunoprecipitation (co‐IP), followed by anti‐βTrCP1 and anti‐ATF4 immunoblot. (G) Expression of ATF4 in SGC‐R (left) or BGC‐R (right) with βTrCP1 knockdown was determined by western blotting. (H) Expression of ATF4 in SGC‐R (left) or BGC‐R (right) with Cullin1 knockdown was detected by western blotting
