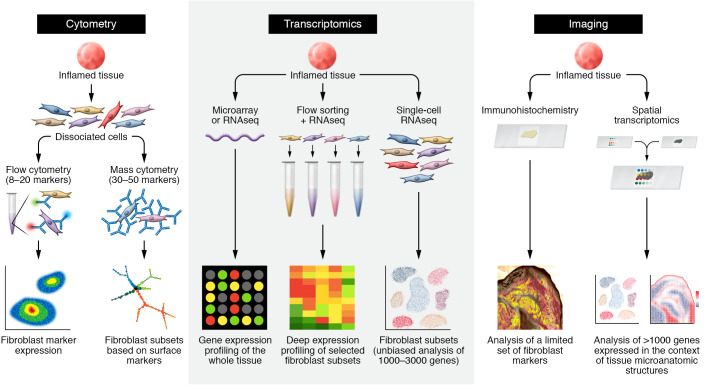
Figure 1. Approaches to examining fibroblast heterogeneity.
Schematics for studies using cytometry (left), transcriptomic (middle), and imaging (right) techniques, illustrating fibroblast heterogeneity in patient-derived tissues. Left: cytometric analysis of fibroblasts starts with staining of cells from dissociated tissues, followed by antibody staining against surface proteins expressed by fibroblasts. Whereas traditional flow cytometry discerns fibroblasts based on several surface markers — typically three to seven surface protein markers — mass cytometry studies can leverage 30 to 50 surface protein markers. In both approaches, putative fibroblast subsets can be nominated by differential expression of surface markers. Middle: transcriptomic analysis. Bulk transcriptomic analysis by microarray or RNA-Seq measures RNA from whole tissue, but does not discern cellular sources of gene expression. Purification of fibroblast or fibroblast subsets from the tissue by FACS or other cell-purification techniques followed by RNA-Seq enables deep expression profiling of selected fibroblast subsets. Single-cell RNA-Seq obviates the need to identify tissue fibroblasts by specific markers through gene expression profiling across diverse cell populations. Fibroblast subsets and states are determined through unbiased analysis of differentially expressed genes at a single-cell level. Imaging analysis: traditional imaging techniques, such as immunohistochemistry and RNA in situ hybridization, when applied to tissues, are limited to examining a few proteins or genes of interest. Spatial transcriptomic techniques capture gene expression profiles from intact tissue sections, enabling simultaneous gene expression profiling in the context of tissue architecture.