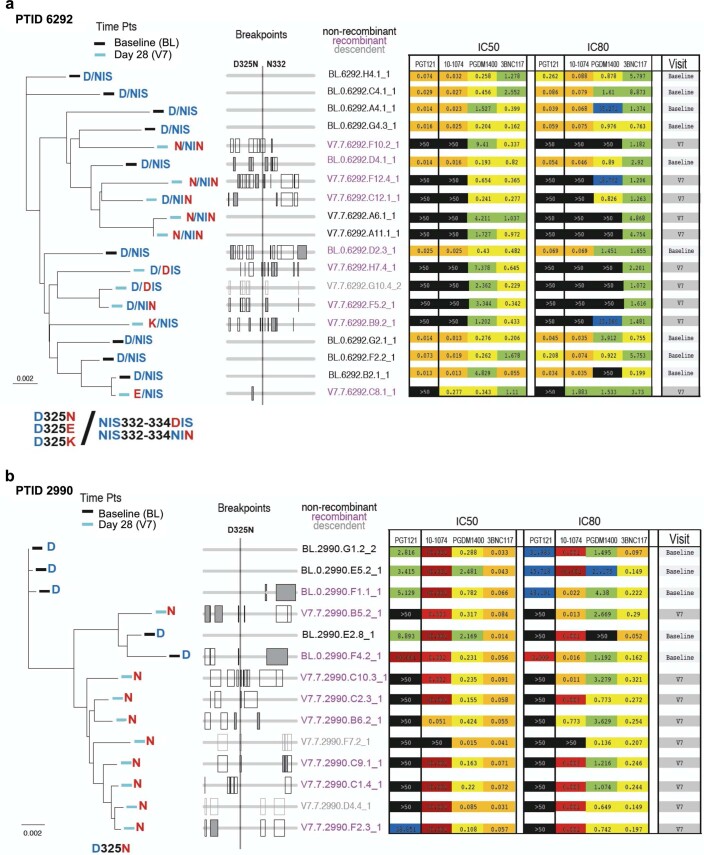
Extended Data Fig. 7. Complex recombination patterns among two participants.
The role of recombination in carrying forward resistant form is shown for study participants 6292 (top) and 2990 (bottom). Left panels show the phylogenetic trees with leaves labeled according to the sensitive/resistant form they are carrying; in the middle panels the corresponding sequences (in the same order as the trees on the left) are each represented by a line, and recombinants display regions where recombination breakpoints likely occurred as rectangles (solid rectangles indicate statistically significant recombination breakpoints); corresponding neutralization data (IC50 and IC80) is shown on the right. In both study participants recombination has no impact at baseline as all forms are PGT121 sensitive. However, complex forms of resistance emerge at visit 7. In 6292, these involve D325 alone, N332 alone, and a combination of both. At visit 7, in 2 out of 2 recombination events between a sensitive and a resistant parental form, the resistant form is found in the recombinant, yet all visit 7 viruses are resistant. Either these mutations are occurring de novo on recombinant backbones, or they are resulting from recombination of parental forms that carry the mutation but have not been sampled. In 2990, recombination carries forward PGT121 resistant forms in 3 out of 3 events where the parental strains carry a sensitive and resistant form each. Of note, D325N does not confer 10–1074 resistance in 2990, neither does D325E in 6292. However, D325K, found in 6292, gives rise to resistance to both 10–1074 and PGT121.