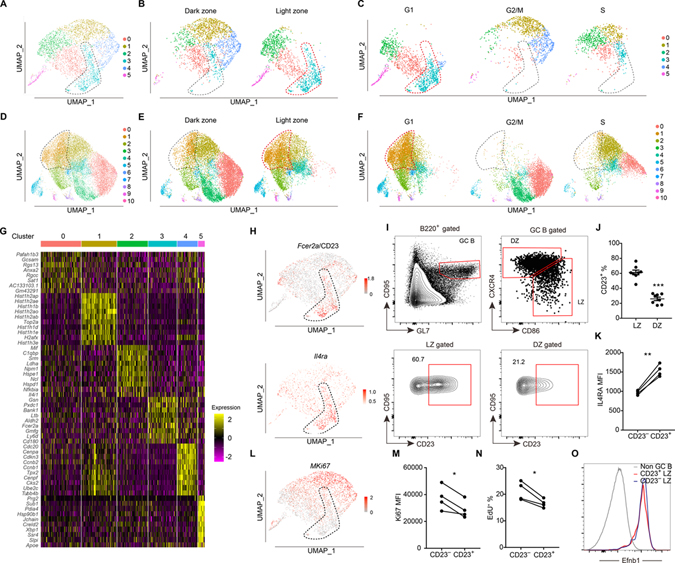
Figure 1. Identification of a CD23+ LZ GC B cell subset by scRNA-seq.
(A-C) UMAP plots of sorted B220+ CD38− GL7hi IgD− GC B cells at day 7 post-immunization (A), split between LZ and DZ (B) and split among different cell cycle phases (C). (D-F) UMAP plots of sorted B220+ CD95hi GL7hi IgD− GC B cells at day 14 post-immunization (D), split between LZ and DZ (E) and split among different cell cycle phases (F). Each point is a single cell colored by cluster assignment. (G) Top 10 marker genes distinguishing the different clusters of GC cells detected at day 7. (H) Expression of Fcer2a (CD23) and Il4ra in the day 7 dataset projected onto UMAP plots. Color scaled for each gene with log normalized expression level. (I, J) Representative FACS profiles (I) and frequencies of CD23+ subset (J) in LZ (CXCR4lo CD86hi) and DZ (CXCR4hi CD86lo) GC B cells 12–14 days after immunization. Each symbol represents one mouse. Data are pooled from two experiments. (K) Geometric mean fluorescence intensity (MFI) of IL4Ra in CD23− and CD23+ LZ GC B cells at day 13 following NP-KLH immunization. (L) Expression of Mki67 from day 7 GC B cell dataset projected onto UMAP plots. (M) MFI of Ki67 in CD23− and CD23+ LZ GC B cells at day 13 following NP-KLH immunization. One of two experiments with similar result is shown. (N) Frequencies of EdU+ cells in CD23+ and CD23− LZ GC B cells after 1 hour EdU labeling. In K, M, N each symbol represents one mouse, each line represents the same mouse, and one of two experiments with similar results is shown. (O) Representative histograms of Efnb1 (Ephrin B1) in non-GC B cells (B220+ CD95lo GL7lo), CD23+ and CD23− LZ GC B cells 12–14 days after immunization. Data represent three experiments, with at least 3 mice per experiment. See also Figure S1 and Table S1.
In this and all subsequent figures, data are presented as mean ± SEM; n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001.