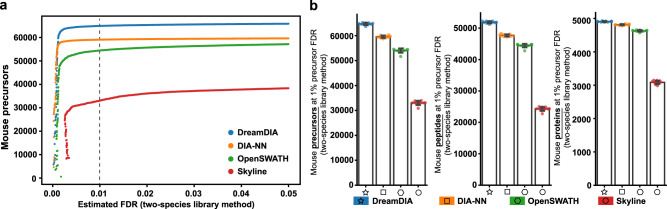
Fig. 2. Identification performance evaluation of DreamDIA on the mouse cerebellum dataset with two-species library method.
a Identification performance on the S1-1 run of the MCB dataset. The numbers of mouse precursors identified at different FDRs were plotted. Each point stands for an Arabidopsis (false-positive) precursor and its discriminant score as a cut-off value. The x-axis value stands for the estimated FDR, calculated as the number of Arabidopsis precursor with higher discriminant score than this cut-off value divided by the number of all the precursors with higher discriminant score than this cut-off value. The y-axis value stands for the number of mouse precursors with higher discriminant score than this cut-off value. b Identification performance on all 10 samples of the MCB dataset. The numbers of mouse precursors, peptides, and proteins at 1% precursor FDR (the respective numbers indicated by the dashed line in (a)) were plotted. Each error bar stands for the mean and standard deviation of the results of n = 10 biologically independent runs.