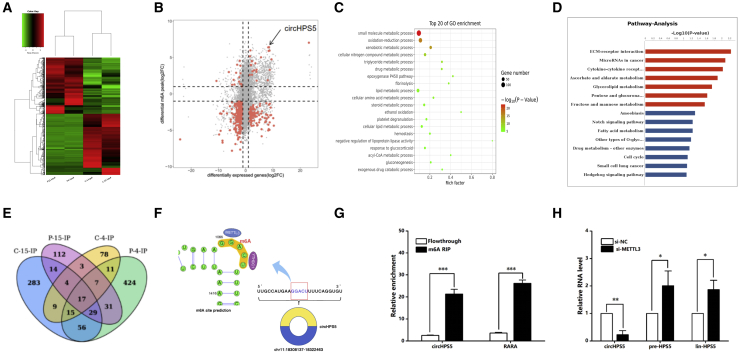
Figure 5.
METTL3 promotes m6A modification and expression of circHPS5
(A) m6A levels were detected in two pairs of HCC and adjacent normal tissues via methylated RNA immunoprecipitation (MeRIP) sequencing. “C” represents “cancer,” and “P” represents “Normal tissue adjacent to cancer.” (B) The association between genes with m6A modification and RNA gene expression levels was analyzed. There were four types of association analysis results: hypermethylated-down (hypermethylated-RNA level down); hypermethylated-up (hypermethylated-RNA level up); hypomethylated-down (demethylated-RNA level down); and hypomethylated-up (demethylated-RNA level upregulated). (C) and (D) GO and KEGG analyses of circRNAs with different m6A modification levels in the HCC group and normal group. (E) The m6A-modified circRNAs in four tissues were cross-analyzed. (F) The SRAMP prediction website revealed the m6A motif structure of circHPS5. (G) MeRIP assays showing the association between circHPS5 and m6A. (H) After knockdown of the methylase METTL3, the expression of circHPS5, pre-HPS5 and lin-HPS5 was detected via qRT–PCR. ∗∗p < 0.01, ∗∗∗p < 0.001.