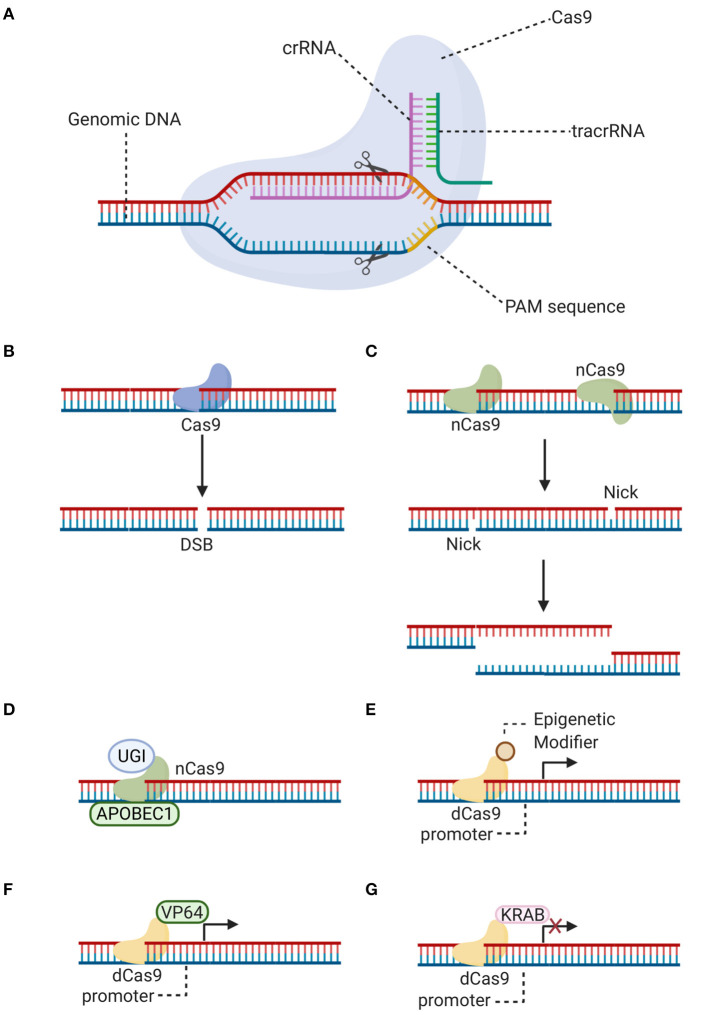
Figure 1.
CRISPR-Cas9 editing. (A) Schematic representation showing how a double-strand break is generated with the CRISPR-Cas9 system. Specificity is brought about by the singue-guide RNA (fused tracrRNA:crRNA), associated with the Cas9 endonuclease, which recognizes its target nucleotide sequence (protospacer). Additionally, there is a requirement for a protospacer adjacent motif [PAM] downstream of the cleavage site. (B) A double-strand break [DSB] can be generated by Cas9 at the target locus, after the single-guide RNA forms a heteroduplex with the protospacer target sequence. (C) Alternatively, a double-strand break can be generated by two Cas9 nickases [nCas9], which are mutant variants that create a “nick” on single target DNA strands. Instead of blunt ends, long overhangs are produced at each of the cleaved ends (a staggered double-strand break). (D) Cas9 nickase (nCas9) is fused to cytidine deaminase (APOBEC1) and uracil glycosylase inhibitor (UGI) for base editing without double-strand breaks. (E–G) Catalytically inactive Cas9 [dCas9] is nuclease-defective but possesses DNA binding ability. dCas9 is fused to epigenetic modifiers such as methyltransferases and acetyltransferases (E), transcriptional activator (VP64) (F) or repressor (KRAB) (G) domains to achieve targeted gene regulation. Illustration created with BioRender software.