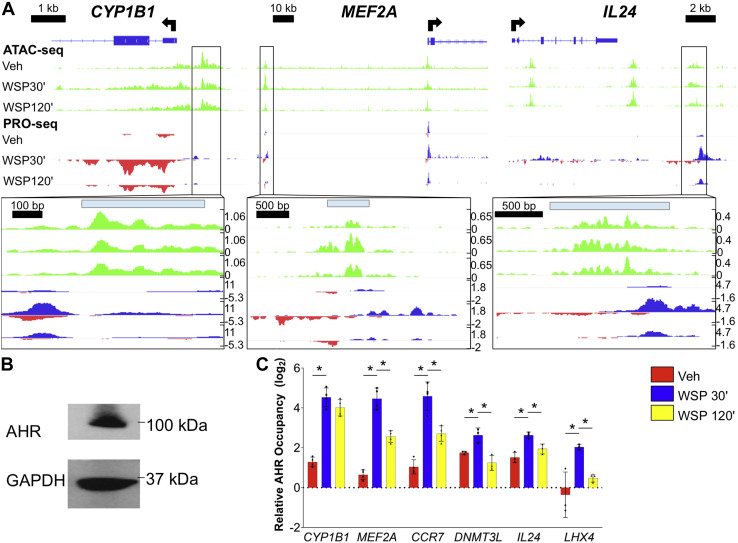
Figure 5.
Integrated analysis of chromatin remodeling and nascent eRNA profiling datasets identifies sites of direct AHR occupancy and functional regulation of early peak transcriptional targets of WSP.A, ATAC-Seq and PRO-Seq tracks aligned in IGV at early peak WSP response genes that are known targets of AHR signaling. Associated early peak eRNAs and ATAC-Seq peaks are boxed and magnified below each panel, with locations of matches to the consensus AHR binding motif, determined using MatInspector, indicated by light blue bars. B, Western blot verification of AHR protein expression in Beas-2B cells under basal culture conditions; GAPDH was a loading control. C, quantitative PCR primers were designed for the three regions examined in (A) plus one additional canonical (CCR7) and two novel (DNMT3L and LHX4) early peak targets of AHR, then ChIP–quantitative PCR was performed in Beas-2B cells following exposure to vehicle or WSP for 30 or 120 min. Bars represent AHR occupancy on a log2 scale (±SD), expressed as the mean CT value at each target region relative to the geometric mean of CT values at three negative control regions (n = 4/group, ∗p < 0.05 for indicated comparison). AHR, aryl hydrocarbon receptor; ATAC-Seq, assay for transposase-accessible chromatin using sequencing; ChIP, chromatin immunoprecipitation; eRNA, enhancer RNA; IGV, Integrative Genomics Viewer; PRO-Seq, precision nuclear run-on sequencing; WSP, wood smoke particle.