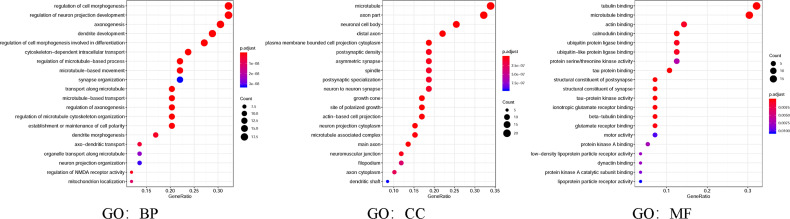
Figure 8.
Gene Ontology (GO) was utilized to analyze the expected functions of MAP mutations and their 50 frequently altered neighbor genes via the DAVID dataset. Starting from the three directions of biological processes (GO: BP), cell components (GO: CC), and molecular functions (GO: MF), the expected functions of target gene mutation can be predicted and analyzed. DAVID, Database for Annotation, Visualization, and Integrated Discovery; MAP, microtubule-associated protein.