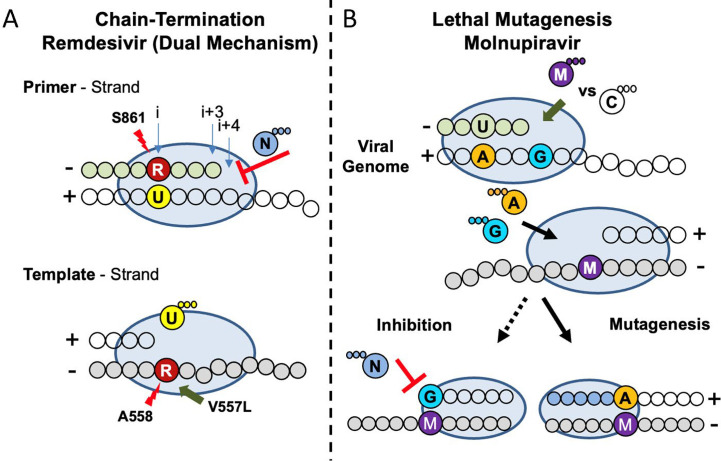
Fig. 8.
Model of the mechanism of action of RDV and molnupiravir against SARS-CoV-2 RdRp. (A) RDV functions as a chain terminator preventing RNA synthesis during both primer and template strand synthesis. (Top) Incorporation of RDV-MP (red) by the viral polymerase (blue oval) opposite UMP in the template (yellow/clear bubbles) results in a steric clash with the residue S861 and incomplete inhibition. (Bottom) The primer is eventually extended and is later used as a template. A clash between RDV-MP and the backbone of A558 is responsible for the reduction in UTP (U) binding. V557L, previously identified in vitro, can compensate for this clash promoting low level resistance. (B) Molnupiravir incorporation results in mutagenesis promoting incorrect base-pairing. (Top) Molnupiravir (purple) primarily base-pairs with GMP (blue), once extended the subsequent template strand promotes ATP (orange) binding opposite molnupiravir explaining lethal mutagenesis observed in cell culture as a result of A-to-G and U-to-C transitions. Biochemical evidence showed evidence of GTP (blue) binding opposite molnupiravir in the template, but this resulted in minimum inhibition and extension of this product would not result in a mismatch.
Adapted from C.J. Gordon, et al., Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template, J. Biol. Chem. (2021) 100770. E.P. Tchesnokov, et al., Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action, J. Biol. Chem. 295 (47) (2020) 16156–16165.