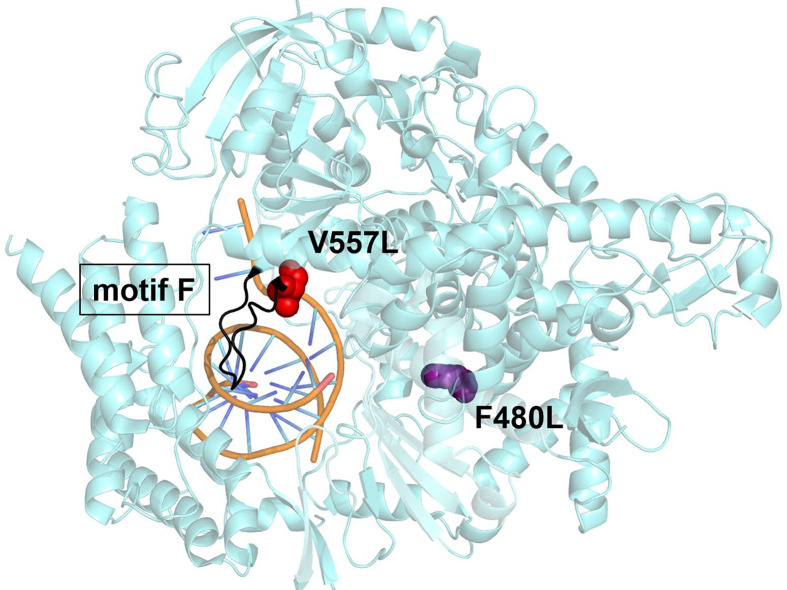
Fig. 9.
Structural representation of the SARS-CoV-2 RdRp (nsp12) containing the resistance conferring mutations, 7BV2. Two RDV resistant mutations selected for in vitro with the coronavirus model murine hepatitis virus (MHV) align to V557L (red) and F480L (purple) in SARS-CoV-2. Both residues are located in the finger subdomain, V557L is associated with structural motif F (black). This motif is implicated with nucleotide incorporation and contacting the 5′ end of the template. Computational and biochemical studies have associated A558 with template-dependent mechanism of RDV, neighboring V557L counteracts this effect resulting in low-level resistance. The distal location of F480L from the active site and V557L suggest it does not have a direct role in RDV resistance as it relates to RNA synthesis. Structures oriented in PyMol, PDB: 7BV2 [208].
Adapted from M.L. Agostini, et al., Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease, mBio 9 (2) (2018) e00221-00218.