FIGURE 5.
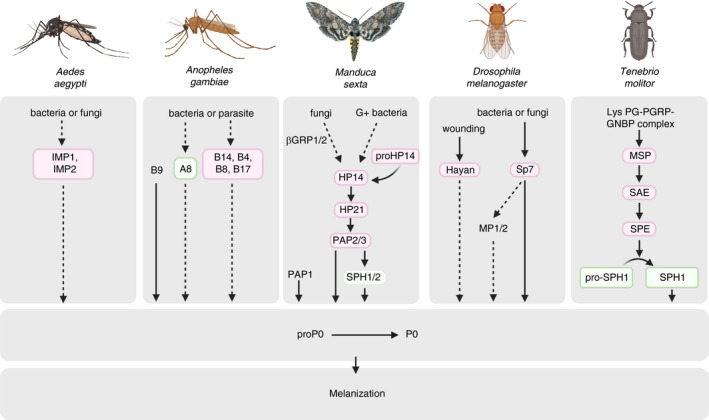
Haemolymph protease cascades in activation of immune‐induced prophenoloxidase (PPO) cascade in different insect species. Upon microbial stimulation, haemolymph pattern recognition proteins (PRRs) like β‐1,3‐glucan recognition proteins (βGRP), peptidoglycan recognition proteins (PGRPs), and c‐type lectins, bind to molecular motifs conserved within surface of microbes, known as pathogen‐associated molecular patterns (PAMPs). This interaction leads to activation of modular serine proteases via a not well‐known mechanism which triggers activation of clip‐domain serine proteases (CLIP proteases) and serine proteinase homologs (SPHs) organized in pathways that eventually lead to activation of PPO to form active phenoloxidase (PO). In Manduca sexta for example, activation of proHP21 by HP14 leads to activation of PPO‐activating proteinase (PAP). SPHs (are also activated by an unknown mechanism) function together with PAPs to form a functional PPO activator to form PO which cleaves PPO to form PO. PO catalyses the oxidation of haemolymph catecholic phenols to corresponding quinones, which can undergo further reactions to form melanin (see Figure 6 for an overall view of this process). For protease names shown in boxes, genetic evidence indicates participation in an immune pathway, but the activating protease and the protease's substrate are not yet known. Proforms (zymogens) of putatively involved proteases are not shown (e.g., M. sexta proHP14 → HP14). Similarly, regulation of proteases by inhibitors known as serpins is not indicted. Arrows indicate activation of downstream components or steps. Steps that have not been verified experimentally are indicated by dashed arrows. Modified after Kanost and Jiang (2015). This Figure summarizes data from various literature, cited parenthetically in the caption of the source diagram (# 2 in reference [247])
