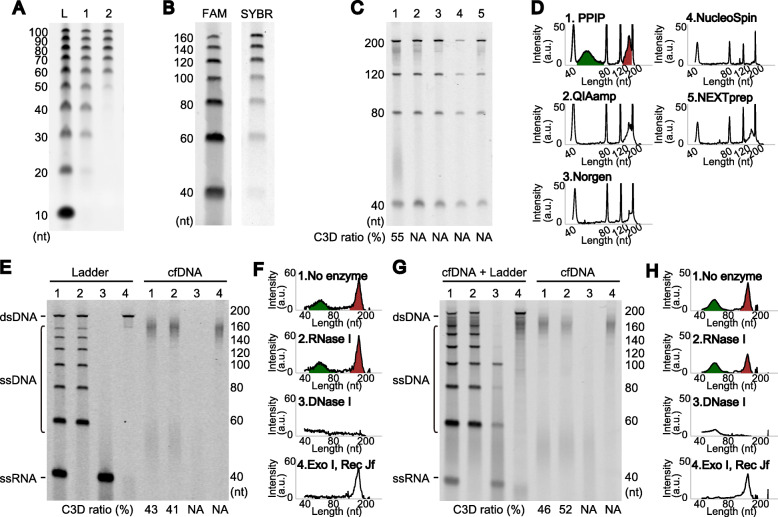
Fig. 1.
Short single-stranded DNA in the cell-free fraction of blood. A Recovery of 10 bp DNA step ladder (lane L, Promega, Fitchburg, WI, USA) was compared using the PPIP method (lane 1) and QIAamp Circulating Nucleic Acid Kit (lane 2). nt; nucleotides. B An ODN mixture was detected with 3′-terminal fluorescent dye labeling (FAM) or staining with SYBR Gold nucleic acid stain (SYBR). C cfDNAs purified from the same plasma using the PPIP scheme or four commercially available kits were compared. The purification methods are PPIP (lane 1), QIAamp Circulating Nucleic Acid Kit from Qiagen (Hilden, Germany) (lane 2), Plasma/Serum Cell-Free Circulating DNA Purification Mini Kit from Norgen Biotek (Thorold, Canada) (lane 3), NucleoSpin Plasma XS from Takara Bio Inc. (Shiga, Japan) (lane 4), and NEXTprep-Mag cfDNA Isolation Kit from PerkinElmer (Waltham, MA, USA) (lane 5). D Electropherograms of the gel images from C. The areas for C3D (green, 40–80 nt) and NPD (red, 120–200 nt) were calculated, and the relative intensity of C3D is indicated. a.u.; arbitrary unit. E–H. Effects of nuclease treatments on a model nucleic acid mixture and cfDNA. The treatment was performed for purified cfDNA (E and F) or in the plasma before purification (G and H). The nucleases used were as follows: no enzyme (lane 1); E. coli ribonuclease I (lane 2); DNase I (lane 3); E. coli exonuclease I and Rec J (lane 4). For the internal control in G, a model DNA mixture was spiked into the plasma (left). For details of the reaction conditions, see Supplementary Methods. Note that the remaining signals in lane 3 disappeared after a long incubation (see Figure S1D). NA denotes “not applicable”