Fig. 4.
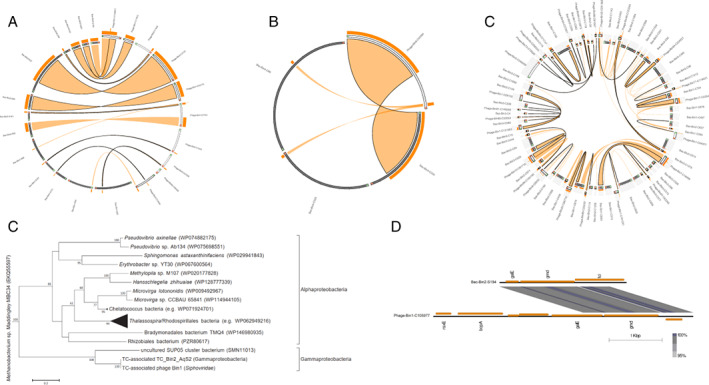
Phage‐host HGT in three vent sponges.
A–C. Circle plots of putatively transferred regions of phages and bacterial hosts in sponge TC (A), sponge HC (B) and sponge HS (C). Viral contigs are shown as protruding grey ideograms; bacterial scaffolds are in black. The direction of orientation is green to red. Orange ribbons show >70% identity. Stacked histograms on top of the ideograms represent the frequency and ratio of the ribbons at each point. Ribbons outlined in black represent the best BLASTn alignment of sequences. Label names have been shortened, for example, Bac‐Bin1‐S36 in (A) represents scaffold_36 of the bacterium TC_Bin1_SUP05; Phage‐Bin52‐C55036 represents contig_55036 of the phage Bin52 in sponge TC.
D. Maximum likelihood‐based phylogeny of galE homologues in TC_Bin2_AqS2 and TC‐associated phage Bin1.
E. Genome structure of contigs containing galE homologues in TC_Bin2_AqS2 and TC associated phage Bin1. Bac‐Bin2‐S194, scaffold_194 of the bacterium TC_Bin2_AqS2; Phage‐Bin1‐C105977, contig_105977 of the phage Bin1. HC, haplosclerid sponge from Crane site; HS, haplosclerid sponge from Swan site; TC, tedaniid sponge from Crane site. HGT, horizontal gene transfer.
