Figure 2.
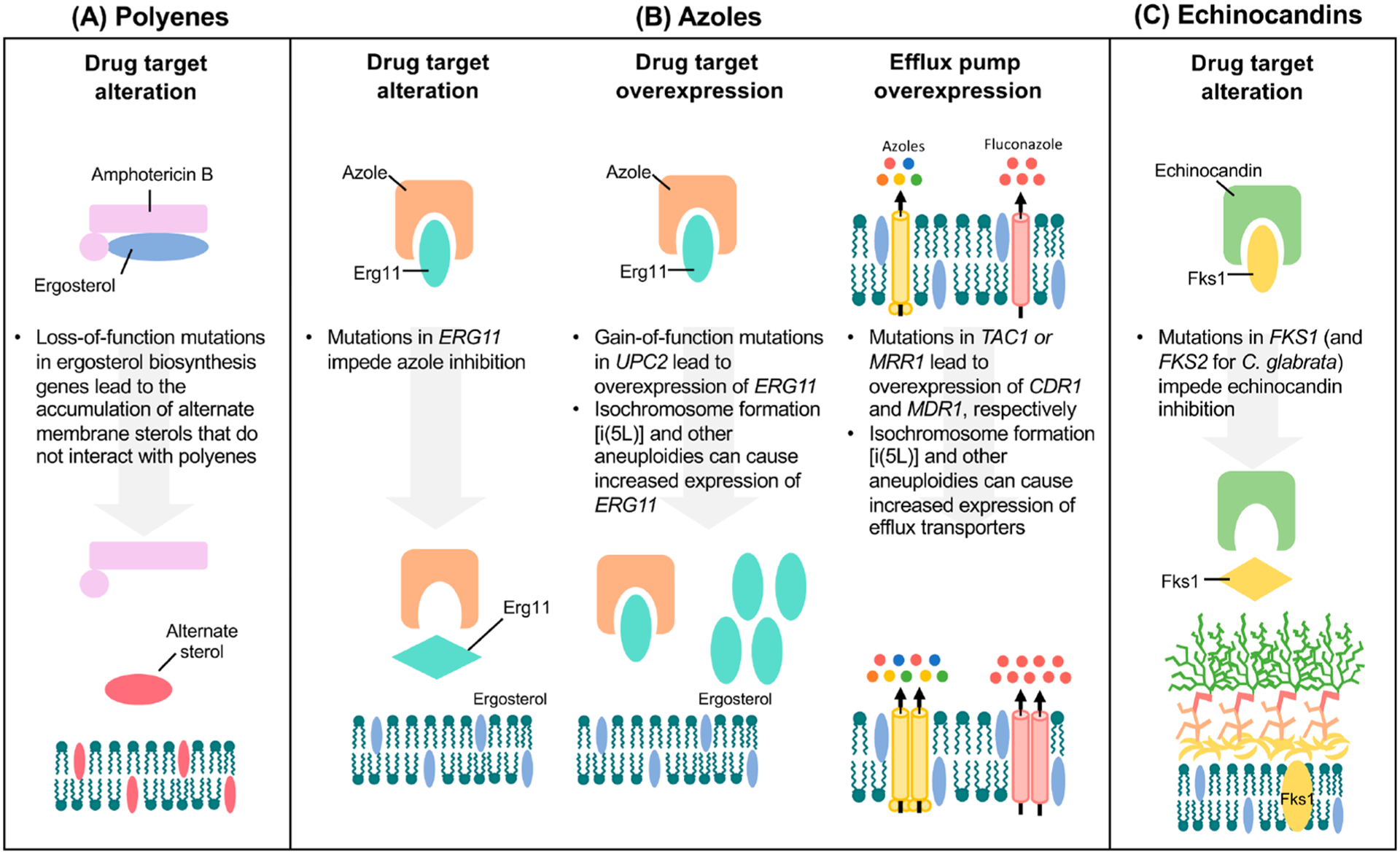
Molecular mechanisms of antifungal drug resistance. (A) Resistance to polyenes is primarily mediated by the depletion of the target ergosterol through loss-of-function mutations in ergosterol biosynthesis genes. This leads to the production of alternate sterols, which do not effectively interact with polyenes and therefore are not extracted from the fungal cell membrane. (B) Resistance to azoles can occur through substitutions to the azole target, Erg11, which leads to lower drug-binding affinity for the lanosterol demethylase enzyme (left panel). Overexpression of the drug target can occur through gain-of-function mutations in the transcriptional activator, UPC2, or through the formation of aneuploidies, such as [i(5L)], which directly increase the copy number of ERG11 (middle panel). Azole resistance is also acquired through the upregulation of ABC transporters (yellow), including Cdr1 and Cdr2, by activating mutations in specific transcription factors (TAC1 in C. albicans and C. auris and PDR1 in C. glabrata). Additionally, overexpression of the MF transporter (pink), Mdr1, through activating mutations in the transcriptional factor, MRR1, confers azole resistance (right panel). Efflux pumps can also be overexpressed through aneuploidy formation. (C) Resistance to echinocandins primarily involves mutations in FKS genes that encode the catalytic subunit of the drug target (1,3)-β-d-glucan synthase. For C. albicans and C. auris, mutations conferring echinocandin resistance occur in FKS1, while for C. glabrata, mutations occur in both FKS1 and its paralogue FKS2.
