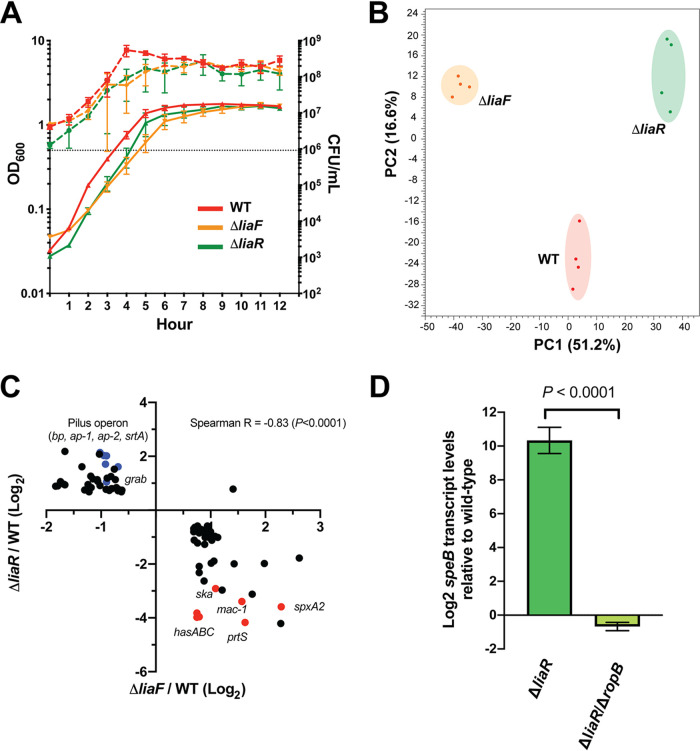
FIG 1.
Transcriptomes of ΔliaF and ΔliaR mutants reveal LiaFSR influence on virulence gene regulation in GAS. (A) Growth phenotypes of parental (WT, red) and isogenic mutant (ΔliaF, orange; ΔliaR, green) strains following growth in vitro in nutrient-rich medium. Growth was performed in biological triplicates and measured by absorbance (OD600, left y axis, solid lines) and CFU (CFU/ml, right y axis, dashed lines). (B) Principal-component analysis (PCA) of parental and isogenic mutant strain transcriptomes (mid-exponential growth in nutrient-rich medium). Biological replicates (colored dots) are indicated for each strain. (C) Correlation plot of 88 significantly (P < 0.05, Bonferroni correction) differentially expressed genes (≥1.5-fold, relative to the parental strain) shared between the ΔliaF (x axis) and ΔliaR (y axis) transcriptomes. Log2 values are plotted, and genes listed in Table 1 are colored (blue or red) and labeled. Red and blue labels indicate genes involved in adherence (blue) or virulence (red). (D) Quantitative real-time PCR comparing speB transcription in the ΔliaR and the ΔliaR ΔropB mutants (relative to the parental strain). Assays were performed in triplicates. P value was determined using the Student’s t test.