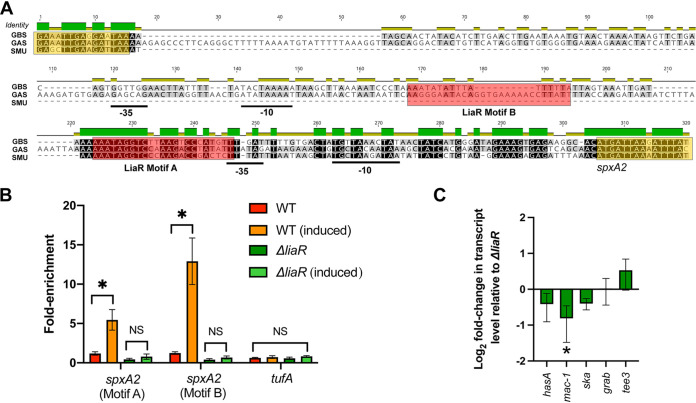
FIG 2.
LiaR directly regulates spxA2 gene expression in GAS. (A) Promoter alignment of spxA2 homologs in GAS (strain MGAS10870), S. mutans ([SMU] strain UA159), and GBS (strain A909). Relative identity is provided above the sequence alignment (green indicates identical). Putative −10 and −35 boxes are underlined. Putative LiaR binding motifs are indicated by red shaded boxes. The 3′ terminal coding sequence for the upstream gene and 5′ coding sequence for spxA2 are indicated by yellow boxes. (B) Chromatin immunoprecipitation (ChIP) quantitative real-time PCR (qPCR) derived from parental (WT) or ΔliaR isogenic mutant strains grown as described in Materials and Methods with or without bacitracin (induced). Primer/probe sequences used in qPCR are provided in Table S4 in the supplemental material. Fold-enrichment relative to the WT (uninduced) is shown on the y axis and the target is indicated on the x axis. *, P < 0.05 (Student’s t test) on assays performed in biological triplicates; comparisons lacking significance are indicated (not significant [NS]). (C) Quantitative real-time PCR comparing target gene transcription of the ΔliaR ΔspxA2 mutant relative to that in the ΔliaR mutant. Shown are transcript levels derived from biological triplicate samples grown in vitro to mid-exponential phase as described in Materials and Methods. *, P < 0.05 (Student’s t test).