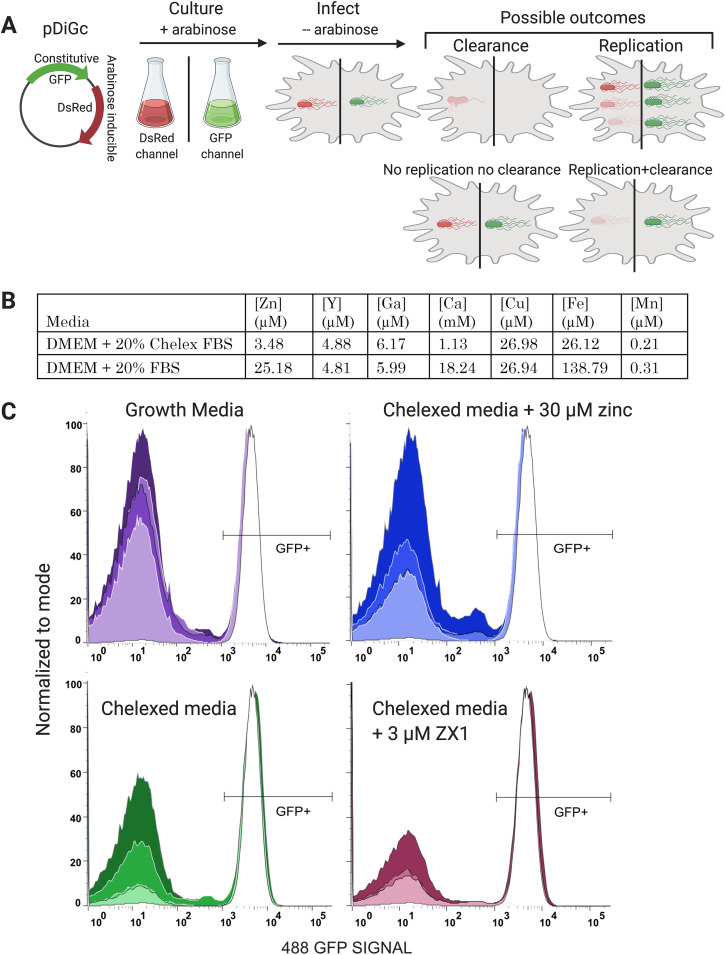
FIG 7.
Bacterial clearance as a function of time and Zn availability. (A) Schematic of the experimental protocol. Salmonella expressing the pDiGc plasmid expresses GFP constitutively and DSRed under arabinose induction. Bacteria are cultured with arabinose and aeration to the stationary phase, at which point they express both fluorescent proteins. Arabinose is removed at infection, and infected macrophages are fluorescent in both red and green channels by flow cytometry. As infection proceeds, the green to red fluorescence ratio “tracks” the bacteria. (B) Metal content of normal growth medium and medium containing Chelex-treated FBS, as determined by inductively coupled plasma mass spectrometry (ICP-MS). (C) Histograms of the GFP signal in macrophages expressing pDiGc in 4 different media and at 4 time points. Each experimental condition was done in triplicate, and 10,000 cells were measured per sample. Replicates were essentially identical, so data for the replicates were concatenated. Increasing color saturation indicates time post Salmonella exposure (2, 10, 18, or 24 h). The right peak in each graph corresponds to the GFP+ macrophages, indicating infection. As Salmonella are cleared, the macrophages become GFP− and shift to the left peak on each graph. At the initial inoculation (white curve in each panel), macrophages show almost 100% infection rate. Normal macrophage growth medium and Chelexed medium supplemented with 30 μM Zn (replete-Zn conditions) show almost 100% bacterial clearance at 24 h. Chelexed medium (low Zn) shows decreased bacterial clearance, while Chelexed medium with 3 μM extracellular zinc chelator ZX1, (essentially no Zn) shows poor bacterial clearance. Figure created with BioRender.com.