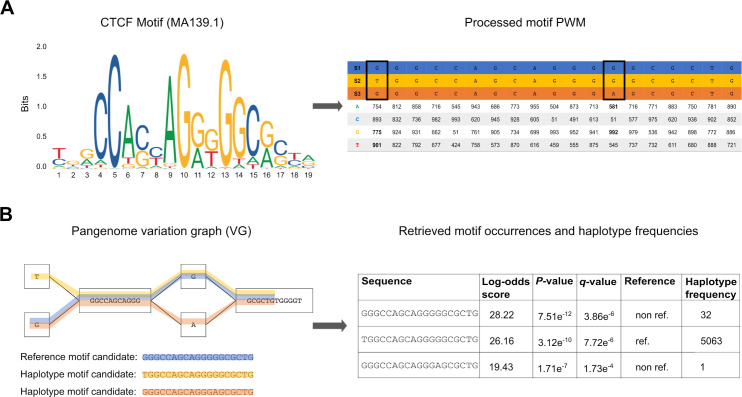
Fig 1. GRAFIMO TF motif search workflow.
(A) The motif PWM (in MEME or JASPAR format) is processed and its values are scaled in the range [0, 1000]. The resulting score matrix is used to assign a score and a corresponding P-value to each motif occurrence candidate. In the final report GRAFIMO returns the corresponding log-odds scores, which are retrieved from the scaled values. (B) GRAFIMO slides a window of length k, where k is the motif width, along the haplotypes (paths in the graph) of the genomes used to build the VG. The resulting sequences are scored using the motif scoring matrix and are statistically tested assigning them the corresponding P-value and q-value. Moreover, for each entry is assigned a flag value stating if it belongs to the reference genome sequence ("ref") or contains genomic variants ("non.ref") and is computed the number of haplotypes in which the sequence appears.