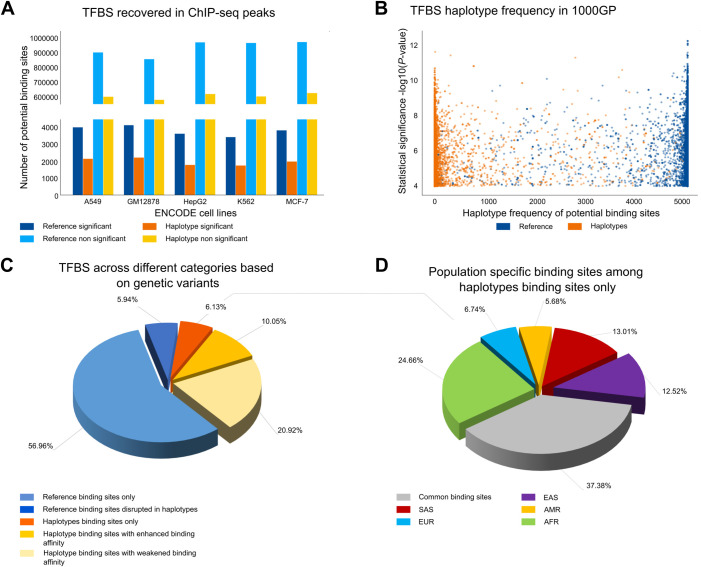
Fig 2. Searching CTCF motif on VG with GRAFIMO provides an insight on how genetic variation affects putative binding sites.
(A) Potential CTCF occurrences statistically significant (P-value < 1e-4) and non-significant found in the reference and in the haplotype sequences found with GRAFIMO oh hg38 1000GP VG. (B) Statistical significance of retrieved potential CTCF motif occurrences and frequency of the corresponding haplotypes embedded in the VG. (C) Percentage of statistically significant CTCF potential binding sites found only in the reference genome or alternative haplotypes and with modulated binding scores based on 1000GP genetic variants (D) Percentage of population specific and common (shared by two or more populations) potential CTCF binding sites present on individual haplotypes.